Artificial antibody produced using partial sequence of enolase protein originated from plasmodium falciparum, and method for producing same
一种序列、抗原的技术,应用在抗体效价检查材料领域,能够解决抗原肽确认试验困难等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0233] (Example 1) Synthesis method of antigenic peptide (I') and antigenic peptide (II') The synthesis diagram is shown below. (E 5 - AD22-PG: serial number 6; E 5 -AD22-PGC: serial number 7; K-E 4 -C: serial number 8; K-E 4 -CK: serial number 9)
[0234] [chemical 8]
[0235] Example of Novel Antigen Threat (I') (AD22map2-SS) Manufacturing Schematic
[0236]
[0237] [chemical 9]
[0238] Example of Novel Antigen Threat (II') (AD22map2pal-SS) Manufacturing Schematic
[0239]
[0240] Initially, monomers of each antigenic peptide (I') and antigenic peptide (II') (AD22map2 and AD22map2pal, respectively) were synthesized according to the usual Fmoc peptide synthesis method. In the Fmoc peptide synthesis method, the resin Fmoc-βAla-PEGresin to which the first amino acid was introduced in advance according to a conventional method was used (the amount of Fmoc amino acid introduced into the resin here is 1 eq). 2% DBU / DMF (or 30% piperidine / DMF) was used in the depro...
Embodiment 2
[0244] (Example 2) Analysis data of antigenic peptide (I') (AD22map2-SS)
[0245] Appearance: white freeze-dried body
[0246] Amino acid analysis value: (hydrolysis condition: 6M HClaq.(with Phenol)110℃, 22hrs)
[0247] Asp(28) 28.00, Thr(8) 7.78, Ser(8) 7.21, Glu(36) 35.62, Gly(4) 4.02, Ala(4) 4.00, Cys(2) 1.36, Leu(4) 4.03, Tyr( 8) 8.00, Lys(10) 10.07, NH3(16) 18.08, Pro(8) 8.18, Phe(8)+β-Ala(2) 10.01.
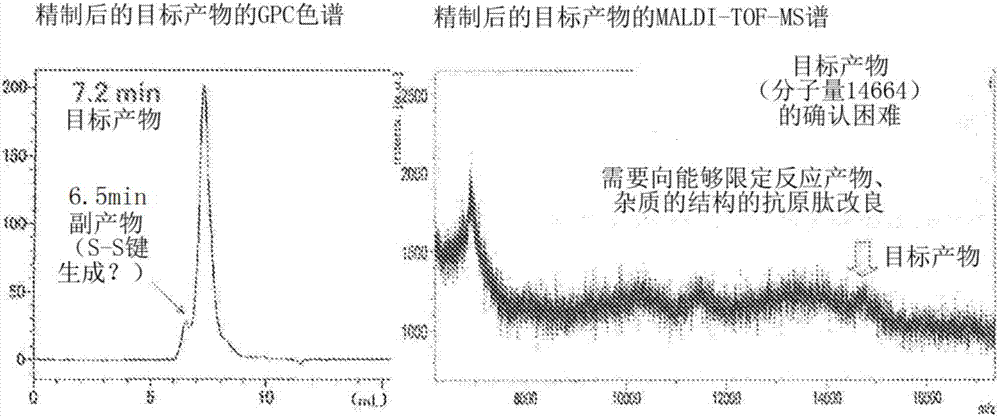
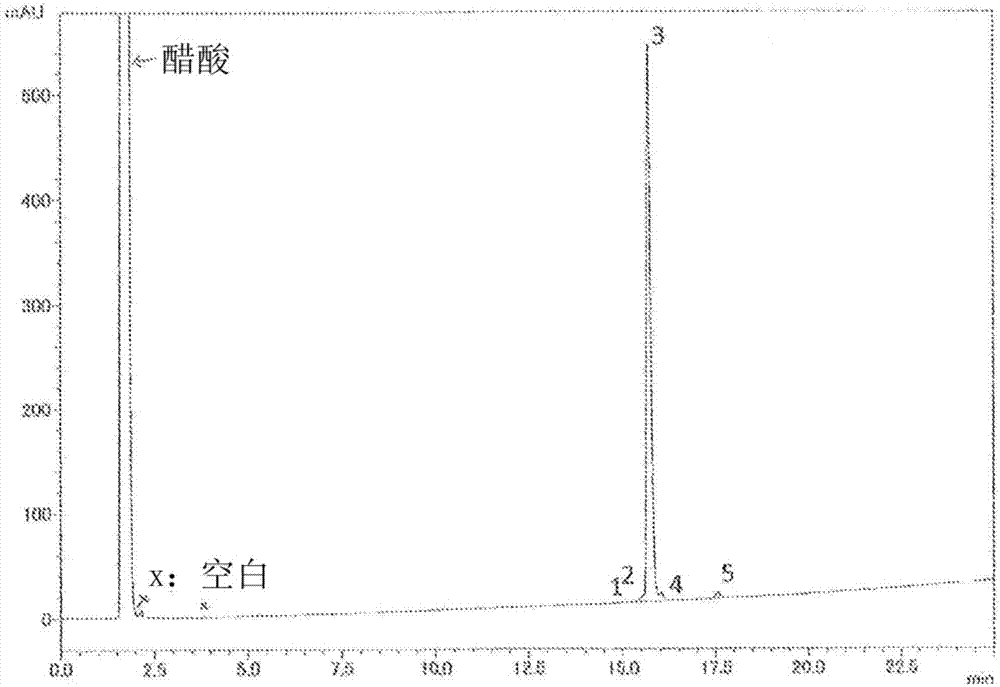
[0248] Purity (HPLC): 96.3% ( image 3 )
[0249] Analysis conditions: column (Column), Zorbax 300SB-C18 (4.6x150mm); eluant (Eluant), 10-60% MeCN / 0.1% TFA (25 minutes); temperature (Temp.) 50 ° C; flow rate (Flow rate ): 1.0ml / min; Detector, 220nm; Load, 4μL (0.28mg / 0.560mL 50% AcOH).
[0250] ESI-MS: MW=15249.3 (theoretical value 15249.3) ( Figure 4 )
[0251] Measuring condition device: HP 1100series LC / MSD manufactured by Agilent Technologies
[0252] Sample concentration: 1nmol / 5micro-L
[0253] Dilution solvent: 50% MeCN / H 2 O: 1N NH 3 aq.=95:5
Embodiment 3
[0254] (Example 3) Analysis data of antigenic peptide (II') (AD22map2pal-SS)
[0255] Appearance: white freeze-dried body
[0256] Amino acid analysis value: (hydrolysis condition: 6M HClaq. (phenol) 110°C, 22hrs)
[0257] Asp(28) 27.96, Thr(8) 7.75, Ser(8) 7.21, Glu(36) 35.70, Gly(4) 4.01, Ala(4) 4.00, Cys(2) 1.90, Leu(4) 4.04, Tyr( 8) 7.81, Lys(12) 11.99, NH3(16) 17.21, Pro(8) 8.20, Phe(8)+β-Ala(2) 9.98
[0258] Purity (HPLC): 96.4% ( Figure 5 )
[0259] Analysis conditions: column (Column), Zorbax 300SB-C18 (4.6x150mm); eluant (Eluant), 30-80% MeCN / 0.1% TFA (25 minutes); temperature (Temp.) 50 ° C; flow rate (Flow rate ), 1.0ml / min; Detector: 220nm; Load, 8μL (0.32mg / 0.320mL 50% AcOH).
[0260] ESI-MS: MW=15982.2 (theoretical value 15982.5) ( Image 6 )
[0261] Measuring condition device: HP 1100series LC / MSD manufactured by Agilent Technologies
[0262] Sample concentration: 1nmol / 5micro-L
[0263] Dilution solvent: 50% MeCN / H 2 O: 1N NH 3 aq.=95:5
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap