Biological marker of epithelial ovarian cancer and application of biological marker
A technology for ovarian cancer and epithelium, which is applied in the field of molecular biology and the preparation of disease diagnostic reagents, and can solve the problems of unknown functional significance of risk variables, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1 patient and sample
[0042] The subjects were 74 patients with familial epithelial ovarian cancer and 47 patients with non-epithelial ovarian cancer (controls). miRNA and mRNA were obtained from lymphocytes in the peripheral blood of the subjects. There were no familial relationships among the 74 patients with familial epithelial ovarian cancer. In the families of these patients, at least two first- or second-degree relatives were diagnosed with genetic ovarian cancer at a certain age. None of these patients had BRCA1 / 2 or MLH1 / MSH2 mutations.
Embodiment 2
[0043] Example 2 Analysis of the target gene relationship and action pathway of epithelial ovarian cancer-related microRNAs and mRNAs studied in the present invention
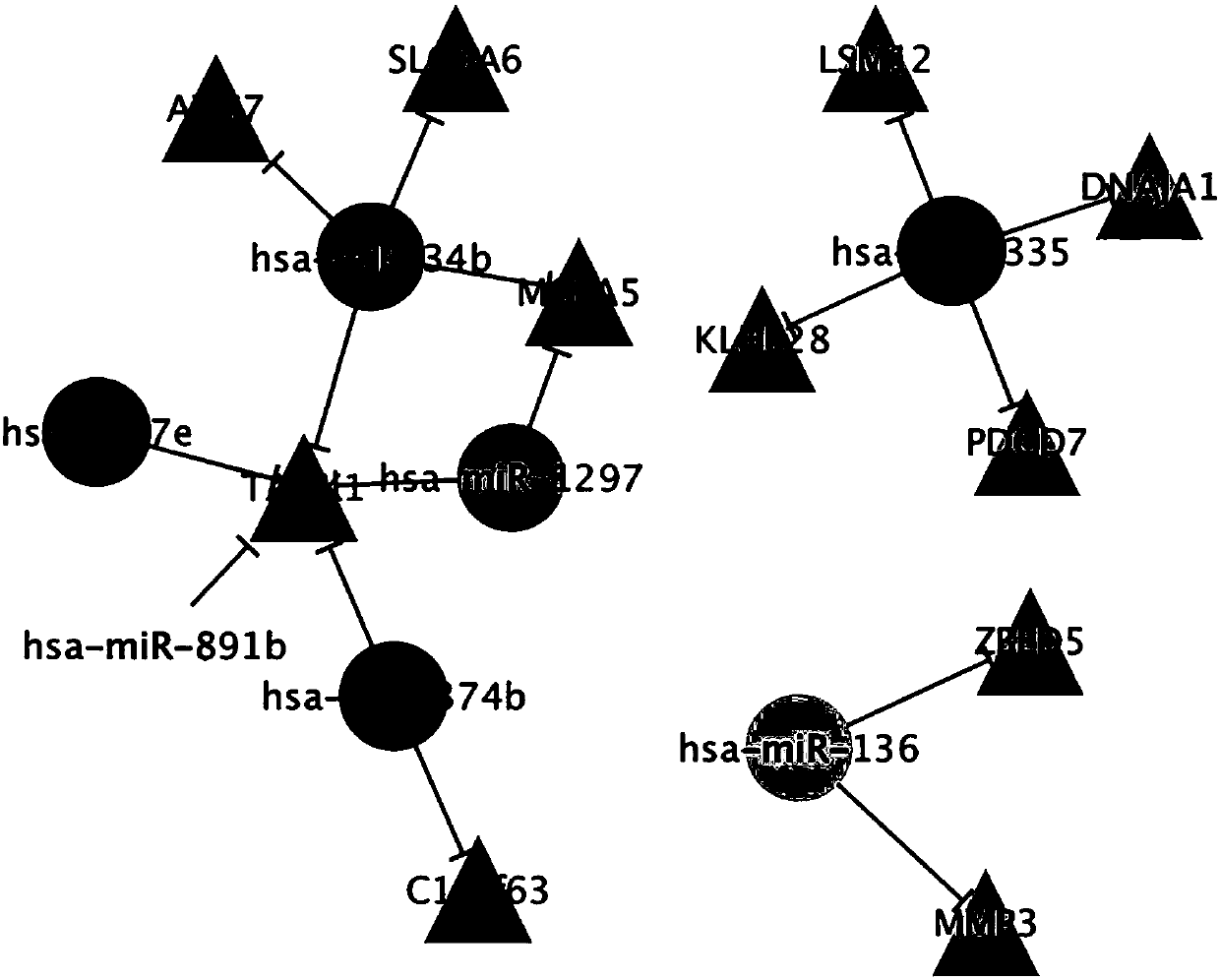
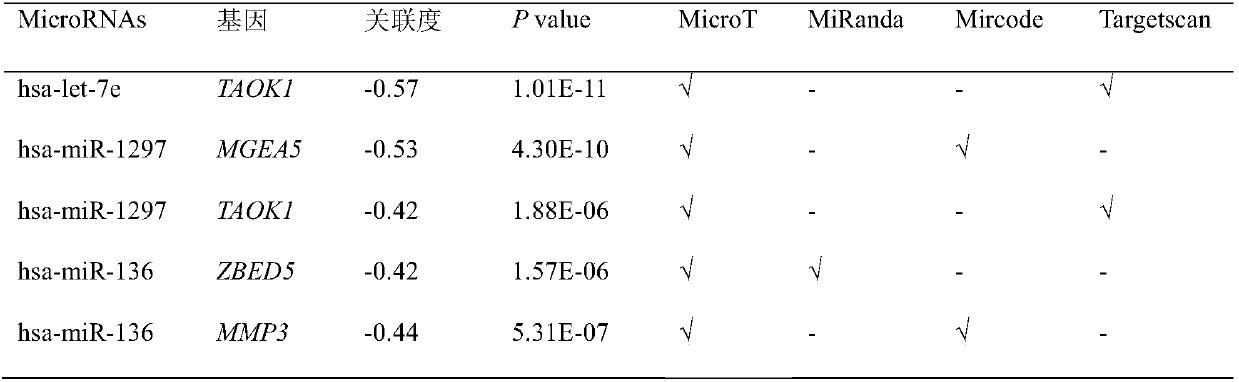
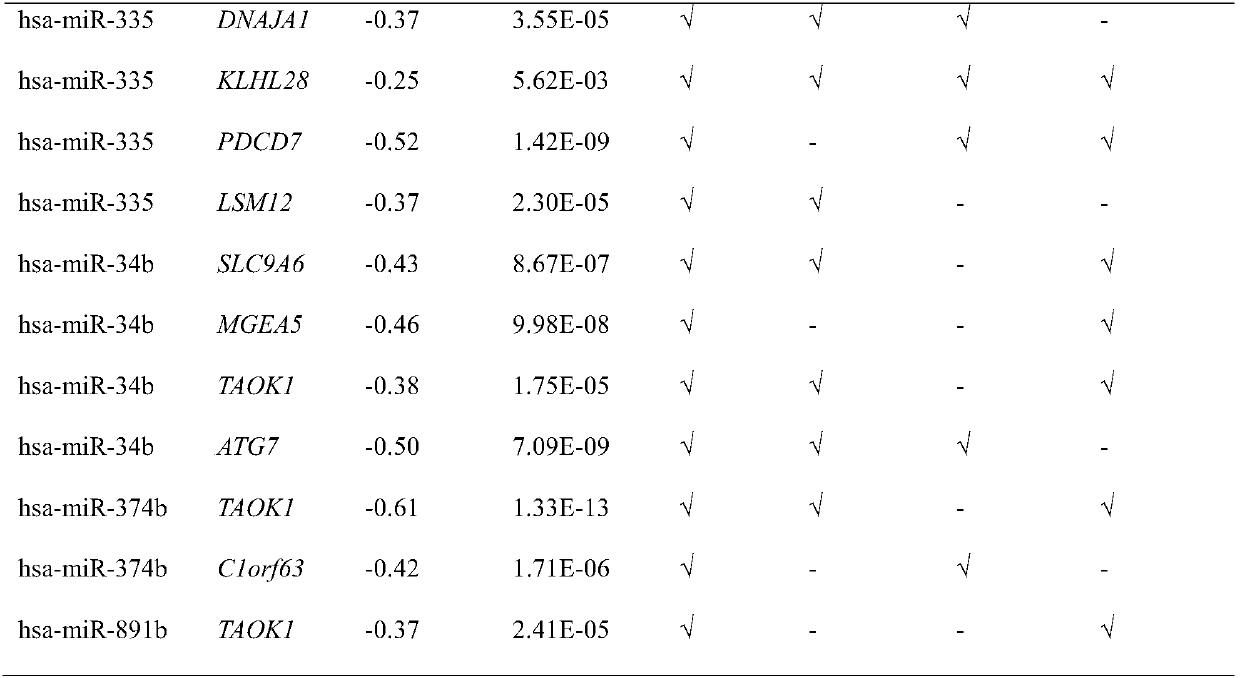
[0044] Using the algorithm shown in Table 1, namely Miranda (microRNA.org), microcode (http: / / www.mircode.org / mircode / ), MirTarget2[21], Targetscan (http: / / www.targetscan.org / ), the microRNAs involved in the method of the present invention, especially the influence factors (microRNA and mRNA) and the action relationship of the core microRNAs (hsa-miR-34b, Hsa-miR-335, hsa-miR-136) are analyzed.
[0045] The results are shown in Table 1. In the samples of epithelial ovarian cancer patients, 16 interacting microRNA and mRNA pairs were found; the involved microRNAs include: hsa-miR-34b, hsa-miR-335, hsa-miR-136, hsa-let-7e, hsa-miR-1297, hsa-miR-374b and hsa-miR-891b; the genes involved include TAOK1, MGEA5, ZBED5, MMP3, DNAJA1, KLHL28, PDCD7, LSM12, SLC9A6, ATG7, C1orf63. In samples from patients with epithelia...
Embodiment 3
[0056] Example 3 gene chip analysis miRNA expression and mRNA expression
[0057] Using the miRNA expression data (data number GSE31801) obtained by the Illumina Human v2 MicroRNA expression beadchip, the gene expression comprehensive database (Gene expression omnibus, GEO) published in: http: / / www.ncbi.nlm.nih.gov / geo / queryacc.cgi?acc=GSE31801) for analysis. The miRNA plate of the chip includes 1146 human miRNAs (>97% miRNAs disclosed in miRBase release 12) to analyze the expression of miRNAs in the aforementioned subjects. Divide the miRNA analysis area of the chip into 3 areas. Among them, the detection of the first region includes hsa-miR-34b, hsa-miR-335, hsa-miR-136; the second region includes hsa-let-7e, hsa-miR-1297, hsa-miR-374b and hsa-miR -891b; the third region includes the remaining miRNAs.
[0058] In addition, the mRNA expression data obtained by the Illumina expression detection chip (llumina HumanHT-12 V3.0 expression beadchip) (data number GSE37582: http...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com