Plant high-efficiency homologous recombination method based on CRISPR/Cas9
A homologous recombination and plant technology, applied in the field of gene editing, can solve the problem of low efficiency of homologous recombination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 PCR amplification of target sequence and vector digestion
[0061] The PCR amplification program used in the construction of the vector in the present invention is as follows: pre-denaturation at 98°C for 2 minutes, 35 PCR cycles (98°C for 10s, 55-60°C for 15s, 72°C for 1-2min). The PCR reaction system is: DNA polymerase ( FastPfu DNA Polymerase (Beijing Quanshijin Biotechnology Co., Ltd.) 1 μL, 10× buffer 5 μL, target template (100ng / μL) 1 μL, upstream and downstream primers (10pM) 1 μL each, and ultrapure water to 50 μL. After the PCR was completed, the PCR product was purified with a PCR purification kit (Beijing Quanshijin Biotechnology Co., Ltd.) for use.
[0062] Vector digestion system: 10U of each required endonuclease (New England Biolabs (NEB) company), 5μL of plasmid (200ng / μL), 1μL of 10×buffer, make up to 10μL with ultrapure water, 37℃ for about 2h, 80℃ Extinguish the fire for 10 minutes and set aside.
[0063] In-fusion cloning (method refer ...
Embodiment 2
[0064] The construction of embodiment 2 pHR04a vector
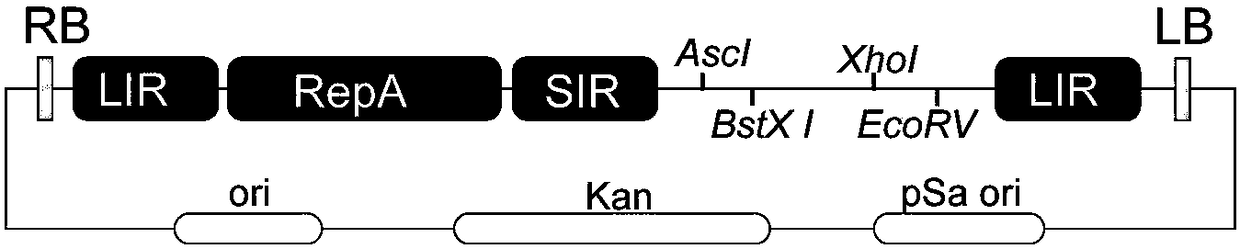
[0065] According to the reference sequence of BeYDV (NCBI NO.DQ458791), its replicon element LIR-RepA-SIR and multiple cloning site (MCS) were synthesized into pGH cloning vector (Shanghai Jierui Biotechnology Co., Ltd.). Firstly, a primer pair was designed (forward: 5-tatatcctgtcaaggcctgagggtcgtacgaataattcgtatccaacggaaatacc-3, reverse: 5-aacgttatcag cttgcatgcgatatcaggtacttttgttctgcga-3). According to the PCR conditions in Example 1, the target fragment LIR-RepA-SIR was amplified, and the pGreen0029 vector was digested with SphI and StuI to construct an intermediate vector. After the sequencing is correct, design primers (forward: 5-aacgttatcagcttgcatgcgagggtcgtacgaataattcgtatccaac-3, reverse: 5-aagtacctgatatcgcatggttgttgtga ctccgagggg-3). Amplify the target fragment LIR according to the PCR conditions in Example 1, cut the intermediate vector with SphI, and clone by In-fusion to obtain the carrier pHR04a( figure 1 ). ...
Embodiment 3
[0066] Example 3 Construction of pHR04a-AsRed Vector
[0067] The expression cassette CasMV35S-AsRed-Nos was recombined between the AscI and BstXI sites of pHR04a. Primer pairs (forward: 5-acgaccctcggcgcgcctgagacttttcaacaaagggtaatatccgga-3, reverse: 5-acgtgacgtacccaaagctctgggatctagtaacatagatgacaccgcgc-3) were designed. Amplify the target fragment CasMV35S-AsRed-TNos according to the PCR conditions in Example 1, cut the pHR04a vector with AscI and BstXI, and obtain the vector pHR04a-AsRed ( figure 2 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com