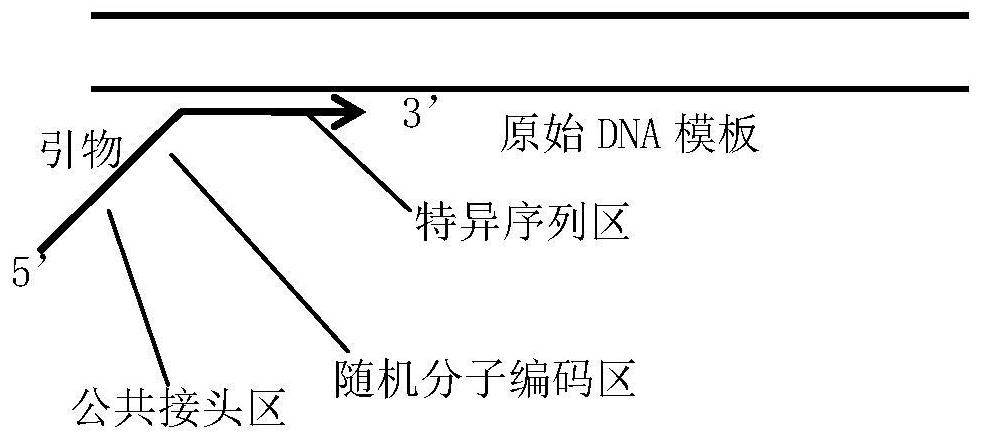
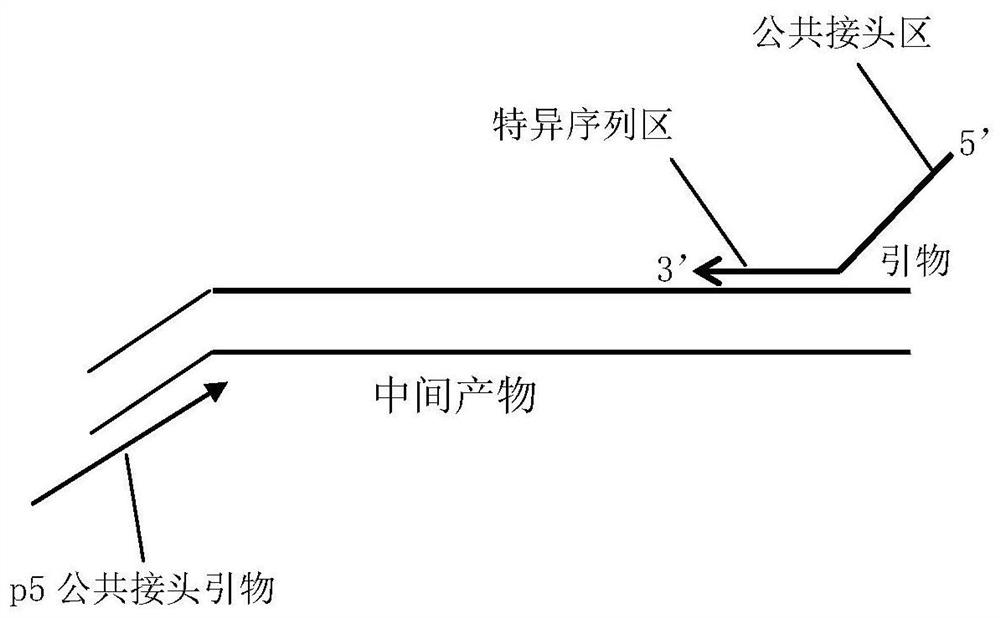
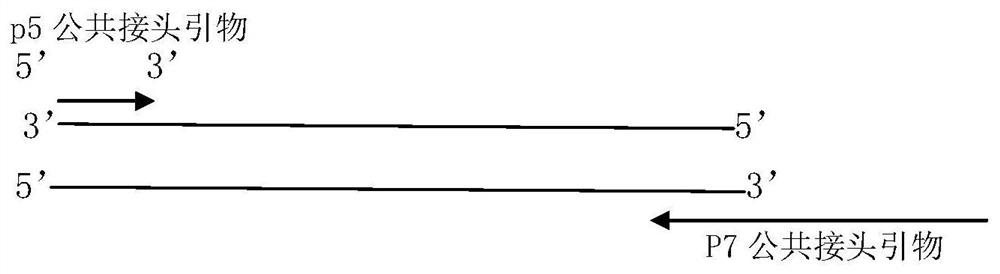
Coding PCR next-generation sequencing library construction method, kit and detection method
A second-generation sequencing and kit technology, applied in biochemical equipment and methods, microbial measurement/testing, chemical libraries, etc., can solve the problems of large amount of DNA, narrow application, insufficient sample volume, etc., to eliminate random errors The effects of matching errors, reducing coding loss, and increasing specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Peripheral Blood Plasma Gene EGFR EXON20 Mutation Detection
[0045] 1. Sample DNA extraction
[0046] Take an appropriate amount of 3-5 ml of peripheral blood plasma from the subject, and use a cell-free DNA extraction kit (DK607-01, provided by Shanghai Laifeng Biotechnology Co., Ltd.) to extract its cell-free genomic DNA.
[0047] 2. Forward single-strand specific linear amplification reaction
[0048] Forward primer EGFR EXON20-55bp-F:
[0049] TACACGACGCTCTTCCGATCTNNNNATTTTTANNNNTAGGAAGCCTACGTGATGGC (SEQ ID NO: 1)
[0050] The forward single-strand specific linear amplification reaction system is: 5×PCR buffer 2 μL, 10mM dNTP 0.5 μL, DNA template 1 μL (containing 24.75ng DNA), 5 μM primer 1 μL, 5U / μl KOD DNA polymerase 0.2 μL, H 2 O 5.8 μL, total volume 10 μL.
[0051] The reaction conditions are shown in Table 1, and a three-step thermal cycle with slow cooling was adopted.
[0052] Table 1 Forward single-strand specific linear amplification reactio...
Embodiment 2
[0096] Example 2 Detection of Multiple Mutations of EGFR EXON18, EGFR EXON19, and EGFR EXON20 in Peripheral Blood Plasma Genes of Tumor Patient Samples
[0097] 1. Sample DNA extraction
[0098] Take an appropriate amount of 3-5 ml of peripheral blood plasma from the subject, and use a cell-free DNA extraction kit (DK607-01, provided by Shanghai Laifeng Biotechnology Co., Ltd.) to extract its cell-free genomic DNA.
[0099] 2. Forward single-strand specific linear amplification reaction
[0100] The forward primer sequence is as follows:
[0101] EGFR EXON20-55bp-F:
[0102] TACACGACGCTCTTCCGATCTNNNNTAAAATNNNNTAGGAAGCCTACGTGATGGC (SEQ ID NO: 7)
[0103] EGFR EXON18-59bp-F:
[0104] TACACGACGCTCTTCCGATCTNNNNTAAAATNNNNGAGATCTTGAAGGAAACTGAATTC (SEQ ID NO: 8)
[0105] EGFR EXON19-58bp-F:
[0106] TACACGACGCTCTTCCGATCTNNNNTAAAATNNNNGAAAGTTAAAATTCCCGTCGCTA (SEQ ID NO: 9)
[0107] Forward single-strand specific linear amplification reaction system: 2 μL of 5×PCR buffer, 0.5 μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com