A nickase and its application in genome base replacement
A technology of cutting enzymes and plant genomes, applied in genetic engineering, enzymes, hydrolytic enzymes, etc., can solve problems such as difficult removal
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
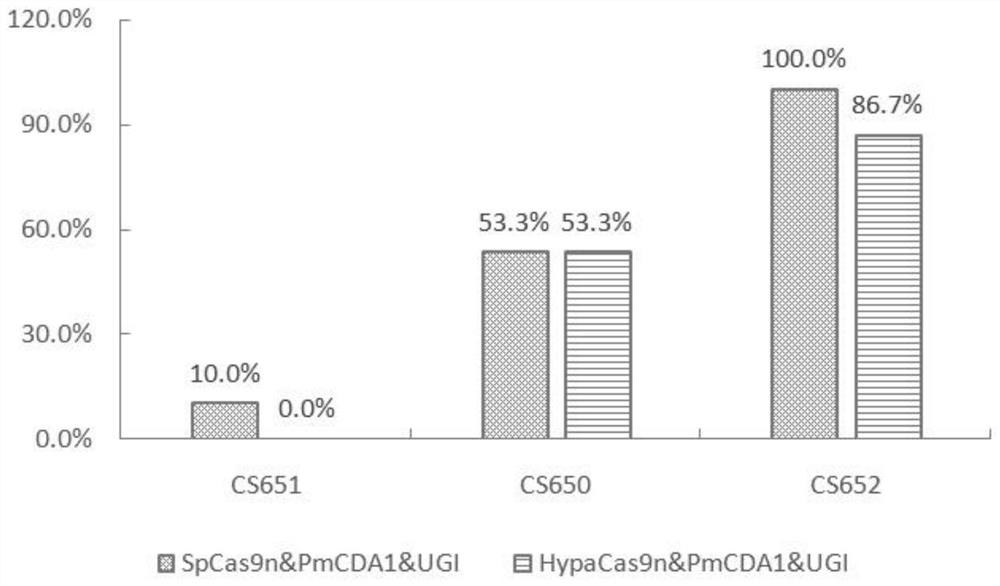
[0035] Example 1, HypaCas9n&PmCDA1&UGI system C T base substitution efficiency
[0036] 1. Construction of genome editing vector
[0037] 1. SpCas9n&PmCDA1&UGI vector: artificially synthesized circular plasmid shown in sequence 1 of the sequence listing.
[0038] Sequence 1 of the sequence listing includes the following three expression cassettes:
[0039] Sequence 1 from the 131st-1283rd position of the 5' end is the expression cassette I, wherein the 131-467th position is the nucleotide sequence of the OsU3 promoter, the 474-550th position is the nucleotide sequence of the pre-tRNA, and the 1st position is the nucleotide sequence of the pre-tRNA. 551-570 is the nucleotide sequence of CS650 target, 571-646 is the nucleotide sequence of sgRNA, 647-723 is the nucleotide sequence of pre-tRNA, 724-743 is the CS651 target The nucleotide sequence of the point, the 744-819th is the nucleotide sequence of the sgRNA, the 820-896th is the nucleotide sequence of the pre-tRNA, and the 89...
Embodiment 2
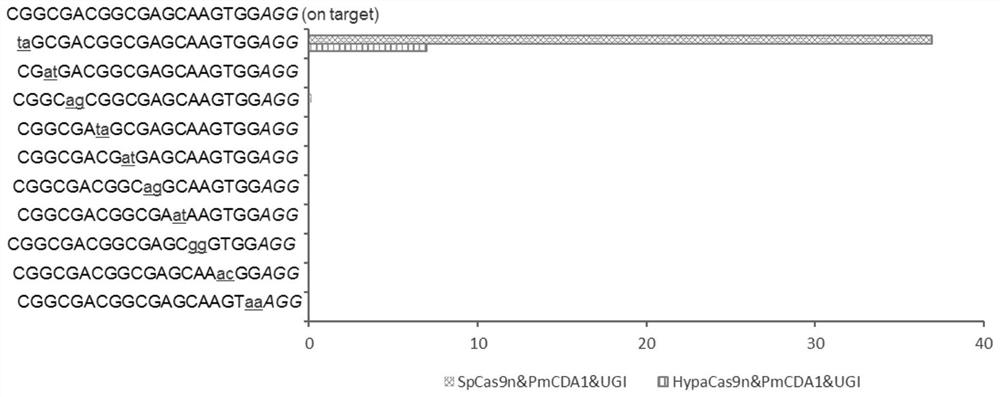
[0062] Example 2, HypaCas9n&PmCDA1&UGI system off-target effect
[0063] 1. Construction of genome editing vector
[0064] SpCas9n&PmCDA1&UGI-T1: A circular plasmid obtained by replacing sequence 1 of the sequence listing from position 551-916 of the 5' end with sequence 3 of the sequence listing.
[0065] SpCas9n&PmCDA1&UGI-T2: A circular plasmid obtained by replacing sequence 1 of the sequence listing with sequence 4 of the sequence listing from 5' end 551-916.
[0066] SpCas9n&PmCDA1&UGI-T3: A circular plasmid obtained by replacing the sequence 1 of the sequence listing from the 551-916 position of the 5' end with the sequence 5 of the sequence listing.
[0067] SpCas9n&PmCDA1&UGI-T4: A circular plasmid obtained by replacing sequence 1 of the sequence listing with sequence 6 of the sequence listing from positions 551-916 at the 5' end.
[0068] SpCas9n&PmCDA1&UGI-T5: A circular plasmid obtained by replacing the sequence 1 of the sequence listing from the 551-916 position ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com