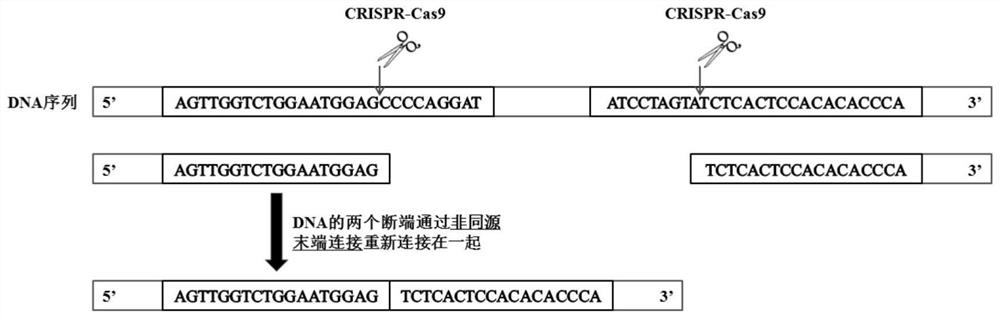
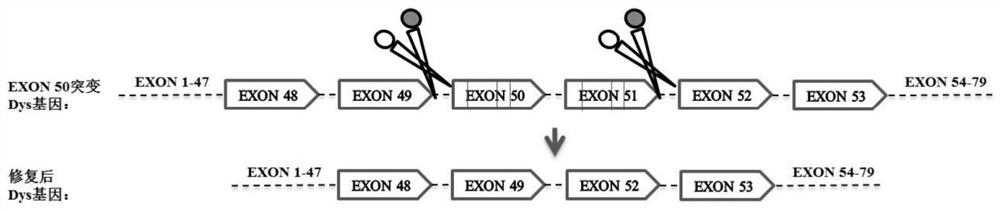
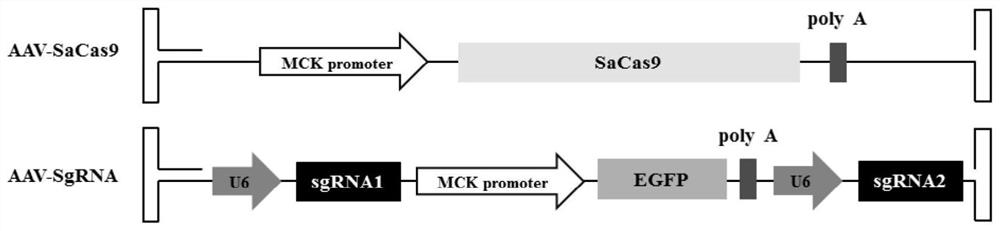
Nucleotide sequence for repairing DMD gene mutation and system
A nucleic acid sequence and gene technology, which is applied in the field of nucleic acid sequence and system for repairing DMD gene mutations, can solve problems such as ineffective packaging, achieve high correction efficiency, improve effective editing efficiency, and low off-target efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1 Screening of gRNA target sequences in introns 49, 50, and 51 of Dys gene
[0066] 1.1 gRNA preparation
[0067] (1) Referring to the Sa.CRISPR / Cas9 target site design rules, design 21nt gRNA target sequences for the sequences of introns 49, 50, and 51 of the Dys gene respectively. The target sequences of the gRNA are shown in Table 1, Table 2, Shown in Table 3;
[0068] (2) Synthesize the sense strand and the antisense strand of the DNA sequence of the gRNA target sequence according to Table 1 (add cacc to the 5'-end of the sense strand, if the first nucleotide at the 5'-end of the sense strand is not guanine G, then Add caccG at the 5'-end of the sense strand; add aaac at the 5'-end of the antisense strand, if the first nucleotide at the 5'-end of the sense strand is not guanine G, then add aaac at the 3'- end plus C);
[0069] (3) The above-mentioned gRNA sense strand and antisense strand were mixed, treated at 90°C, cooled naturally to room temperature f...
Embodiment 2
[0111] Example 2 Sequencing Analysis Dystrophin Gene Mutation Efficiency and Mutation Type
[0112] Select the gRNA with higher mutation efficiency in Example 1, and further analyze the mutation efficiency and mutation type. With SEQ ID NO:10, SEQ ID NO:23 and SEQ ID NO:43; SEQ ID NO:105 and SEQ ID NO:107; SEQ ID NO:152, SEQ ID NO:153, SEQ ID NO:167 and SEQ ID Take NO:179 as an example to perform the following experimental operations.
[0113] 2.1 T carrier connection
[0114] (1) With the purified PCR product obtained in 1.5 (4) of Example 1, measure the concentration for subsequent use;
[0115] (2) Use Kangwei Century Master PCR Mix to add an adenine (A) to the 3'-end of the PCR product: take 2 μg of the PCR product in 3.1 and mix it with the Master PCR Mix at a ratio of 1:1 (V:V), and store at 72°C React for 30 minutes;
[0116] (3) Use the reaction product in 1% agarose gel electrophoresis 4.2, cut the gel to recover the target fragment and measure the concentration o...
Embodiment 3
[0137] Example 3 Screening of gRNA target sequence combinations in introns 49, 50, and 51 of Dys gene
[0138] 3.1 Design of gRNA target sequence combination ( Figure 43 )
[0139] For DMD mutants with deletion of exon 50, a plan to knock out exon 51 can be adopted, and the gRNA target sequences of intron 50 and intron 51 can be selected for combination;
[0140] For DMD mutants with deletion of exon 51, a plan to knock out exon 50 can be adopted, and the gRNA target sequences of intron 49 and intron 50 can be selected for combination;
[0141] For DMD mutants with mutations or deletions in exons 50-51, a plan to simultaneously knock out exons 50 and 51 can be adopted, and the gRNA target sequences of intron 49 and intron 51 can be selected for combination;
[0142] Table 5 Dys gene exon 50-51 upstream and downstream Sa-gRNA combinations
[0143] gRNA for intron 49 gRNA for intron 51 Group NO:1 SEQ ID NO: 23 SEQ ID NO: 153
[0144] Table 6 Sa-gR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com