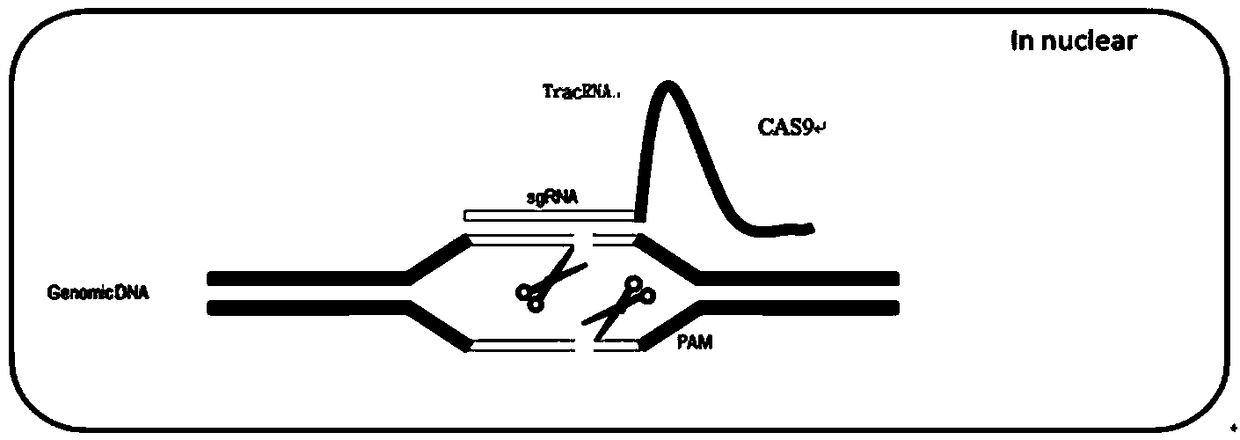
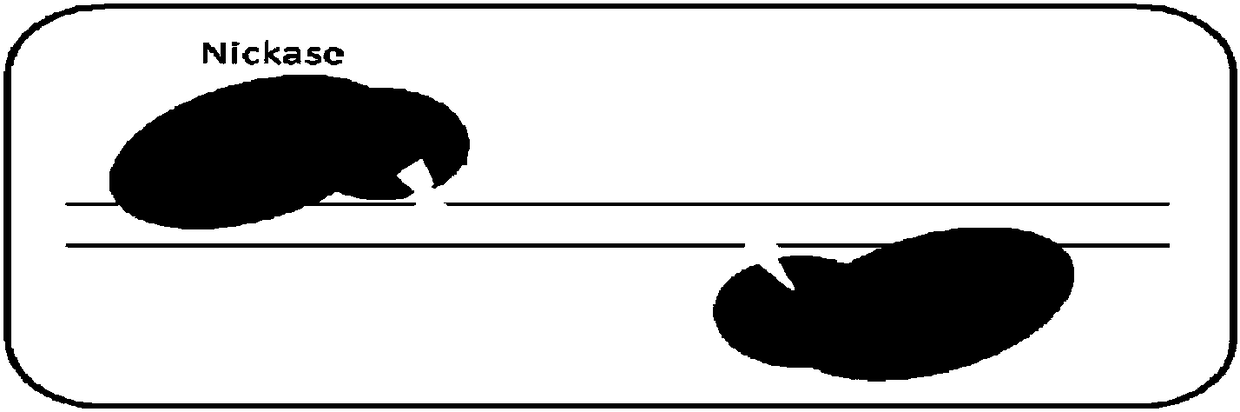
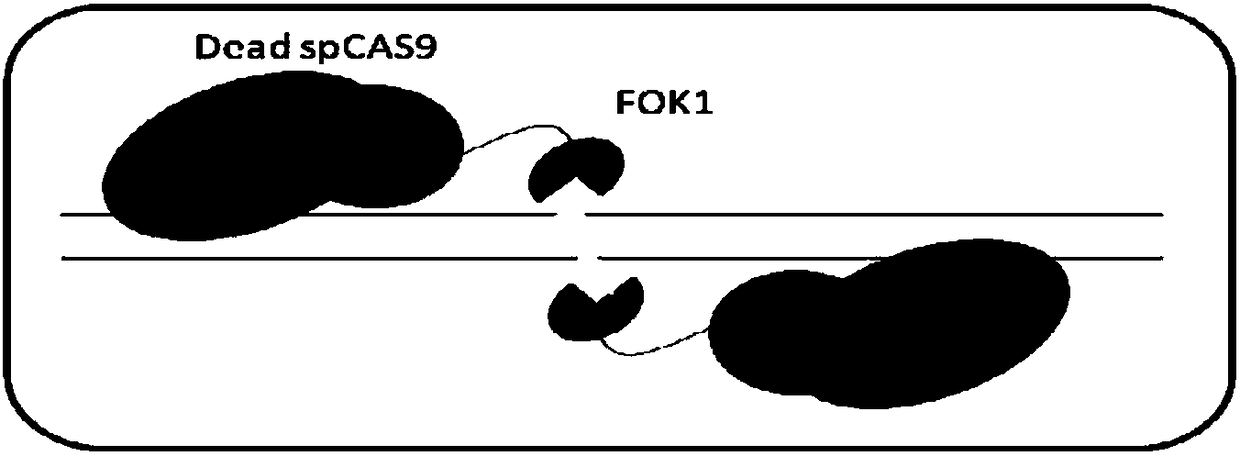
Dead SaCas9-Fok1 system, as well as construction method and application thereof
A deadsacas9-fok1 and construction method technology, applied in the field of gene editing systems, can solve the problems of reducing off-target efficiency, large distance tolerance, short recognition sequence, etc., and achieve the effects of improving targeting efficiency, reducing off-target efficiency, and small molecular weight
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Obtaining of U6-sgRNA expression cassette and construction of U6-sgRNA expression vector
[0042] 1. Synthesis of target fragments
[0043] (1) Synthesize the U6-sgRNA fragment, which is inserted into the Sma I site of the pUC19 vector, wherein the U6-sgRNA fragment is shown in SEQ ID NO: 6, using pUC19-U6-sgRNA as a template, using the primer SaCas9-U6- L / SaCas9-gRNA-R, PCR amplification of the target fragment U6-sgRNA;
[0044] SaCas9-U6-L: 5'-GTACGGGCCAGATATACGCGT-3' (SEQ ID NO: 7)
[0045] SaCas9-gRNA-R: 5′-CAATAATCAATGTCAACGCGT-3′ (SEQ ID NO: 8)
[0046] (2) Purify the above PCR product with a DNA purification kit.
[0047] 2. Expression vector preparation
[0048] (1) The commercial vector pSaCas9-GFP (plasmid map as shown in Figure 6 Shown) Escherichia coli expanded culture, extract plasmid pSaCas9-GFP;
[0049] (2) Digest pSaCas9-GFP plasmid with restriction endonuclease Mlu I-HF, digest at 37°C for 1 hour, and inactivate the enzyme at 65°C for ...
Embodiment 2
[0058] Example 2 Obtaining of dead SaCas9 gene and construction of dead SaCas9 expression vector
[0059] (1) Transformation of pSaCas9nD10A
[0060] 1. Synthesis of target fragments
[0061] (1) Outsourced synthesis of the SaCas9nD10A mutant fragment, which was inserted into the Sma I site of the pUC19 vector;
[0062] The sequence of the SaCas9nD10A mutant fragment is shown in SEQ ID NO: 9;
[0063] (2) Using pUC19-SaCas9nD10A as a template, using primers SaD10A-L / SaD10A-R, PCR amplifies the target fragment SaCas9nD10A;
[0064]SaD10A-L: 5'-CATCATTTTGGCAAAGAATTC-3' (SEQ ID NO: 10)
[0065] SaD10A-R: 5'-GACAGCTTCTGACTCAGGCCT-3' (SEQ ID NO: 11)
[0066] (3) Purify the above PCR product with a DNA purification kit.
[0067] 2. Expression vector preparation
[0068] (1) The pSaCas9-gRNA plasmid obtained in Example 1 was double digested with restriction endonucleases EcoR I and Stu I, digested at 37°C for 1 hour, and inactivated at 65°C for 20 minutes;
[0069] (2) After t...
Embodiment 3
[0095] Example 3 Obtaining of Fok1 gene and construction of dead SaCas9-Fok1 expression vector
[0096] 1. Synthesis of target fragments
[0097] (1) Outsourced synthesis of the Linker-Fok1 fragment, which was inserted into the Sma I site of the pUC19 vector;
[0098] The Linker-Fok1 fragment sequence is shown in SEQ ID NO: 15;
[0099] (2) Using pUC19-Linker-Fok1 as a template, use primers Fok I-EcoN I-L / Fok I-Pst I-R to amplify the target fragment Linker-Fok1 by PCR;
[0100] FokI-EcoNI-L: cgacattctg ggaaacctgt atgaggtgaa gagcaaaaag cac (SEQ ID NO: 16)
[0101] FokI-PstI-R: catgagatccccgcgctgcagttaaaagtttatctcacc (SEQ ID NO: 17);
[0102] (3) Purify the above PCR product with a DNA purification kit.
[0103] 2. Expression vector preparation
[0104] (1) The p-dead SaCas9 plasmid obtained in Example 2 (2) was double-digested with restriction endonucleases EcoN I and Pst I, and after digestion at 37°C for 1 hour, the enzyme was inactivated at 65°C for 20 minutes;
[0105...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com