CRISPR/Cas9 recombinant lentiviral vector containing gRNA sequence specifically targeting ccr5 gene and its application
A technology of recombinant lentivirus and lentiviral vector, which is applied in the field of genetic engineering, can solve the problem of incomplete inhibition of HIV replication and achieve high inhibition and rapid construction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] The features and advantages of the present invention can be further understood through the following detailed description in conjunction with the accompanying drawings. The examples provided are only illustrative of the method of the present invention and do not limit the rest of the present disclosure in any way. [Example 1] Design, synthesis and eukaryotic expression vector construction of gRNA sequence targeting CCR5 gene sequence
[0043] 1. Selection and design of gRNA targeting CCR5 gene sequence
[0044] Using the human CCR5 gene sequence as a reference sequence, use the Crispr design tool on crispr.mit.edu to find the target site with a higher score on the CCR5 gene sequence. The target site sequence is in the form of 5'-(20N)-NGG3' or 5'-CCN(20N)-3'. At the same time, the human genome sequence was compared to exclude gRNAs with high homology to the human genome sequence, and the genome sequences of CCR5 and CCR2 were compared to exclude gRNA sequences targeti...
Embodiment 2
[0062] [Example 2] The gRNA-guided CRISPR / Cas9 system of the present invention was used to verify the mutation of a specific site in the CCR5 gene in TZM-bl cells.
[0063] First, the constructed gRAN / Cas9 co-expression plasmid and the helper plasmids psPAX2 and pMD2.G were co-transfected into 293T cells. After 24 hours, the culture medium was replaced with fresh medium. After 48 hours, the culture supernatant was collected, filtered through a 0.45 μm filter membrane, and then cryopreserved. Standby at -80°C.
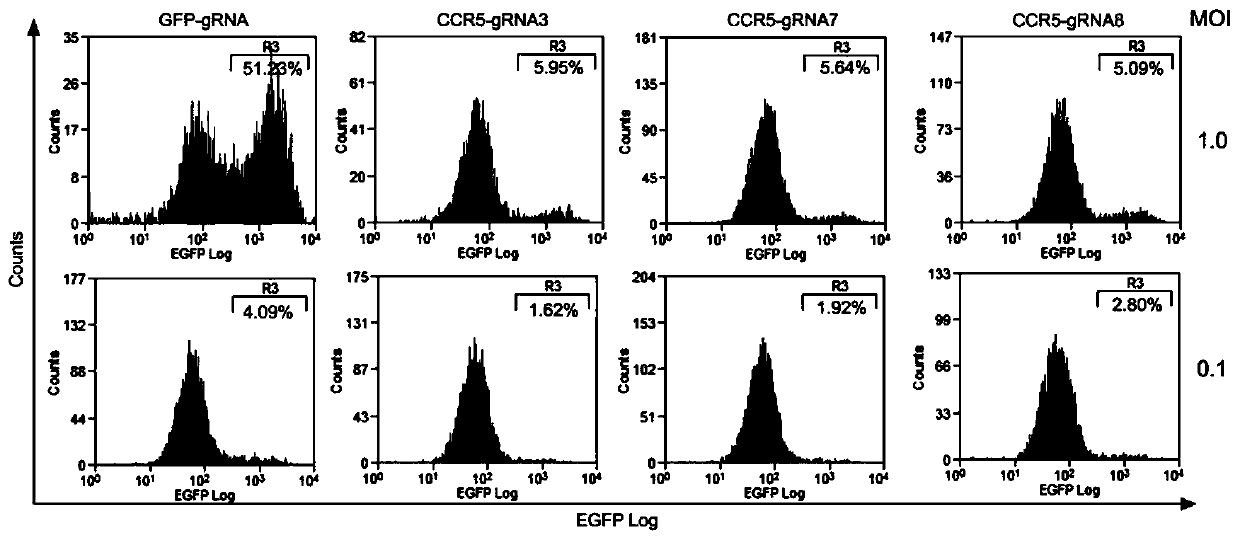
[0064] In order to verify whether the gRNA-guided CRISPR / Cas9 system designed in the present invention can mutate the specific site of CCR5, TZM-bl cells were infected with the constructed lentivirus, replaced with fresh medium after 12 hours of infection, and added 0.8 μg / ml of puromycin kills uninfected cells, the solution is changed every 24 hours, and the genomic DNA of the cells is extracted after 72 hours. Use the CCR5 primer CCR5-Primer to amplify the target fra...
Embodiment 3
[0095] [Example 3] Inhibition of CCR5 co-receptor expression by designed and constructed gRNA
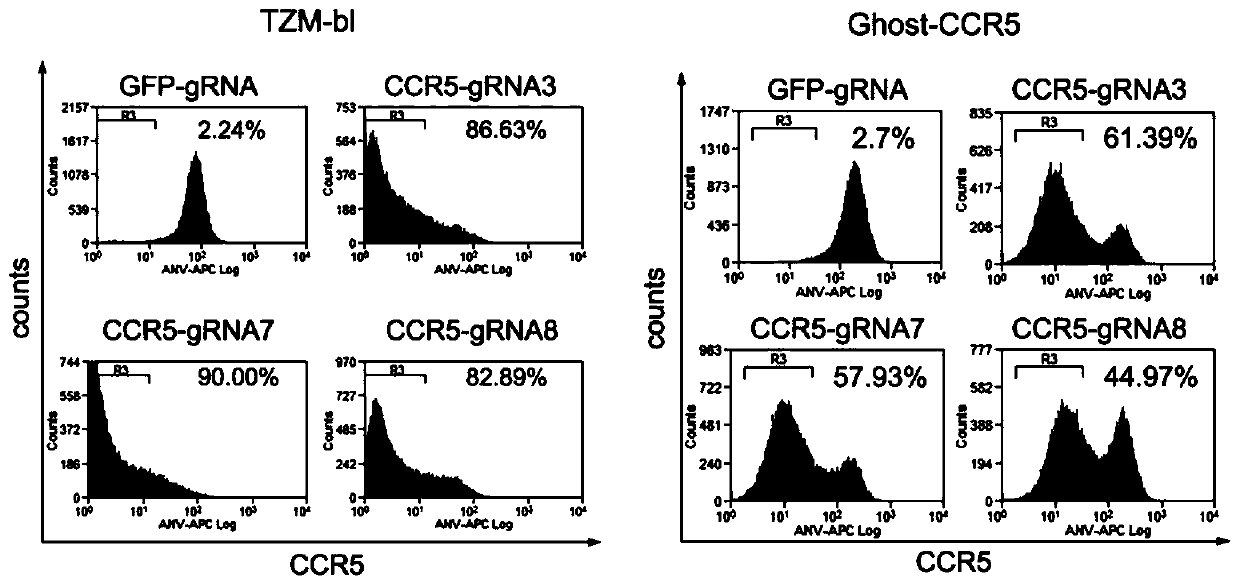
[0096] The virus infection experiment was the same as in Example 2. After 72 hours, the expression of the CCR5 co-receptor on the surface of TZM-bl cells and Ghost-CCR5 cells was detected by flow cytometry, from figure 2 It can be seen that the CRISPR / Cas9 system can effectively inhibit the expression of CCR5 coreceptor under the guidance of our designed CCR5-gRNA7 and CCR5-gRNA8, especially in TZM-bl cells, the specific steps are as follows:
[0097] a. Discard the medium and wash the cells once with PBS;
[0098] b. Add 200 μl of 0.25% trypsin to digest the cells, and then add 300 μl of serum-containing medium to terminate the trypsinization;
[0099] c. Add PBS to 1ml, pipette the cells repeatedly until they are dispersed into single cells;
[0100] d. Transfer the cell suspension to a 1.5ml EP tube, centrifuge to pellet the cells (1,500g, 5min); wash 1-2 times with PBS, repea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com