A CRISPR-Cas9 system for simultaneously knocking out Kras gene and EGFR gene and its application
A gene and DNA sequence technology, applied in the field of CRISPR-Cas9 system, to achieve the effects of simple operation, high knockout efficiency and multiple options
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] 1. sgRNA design
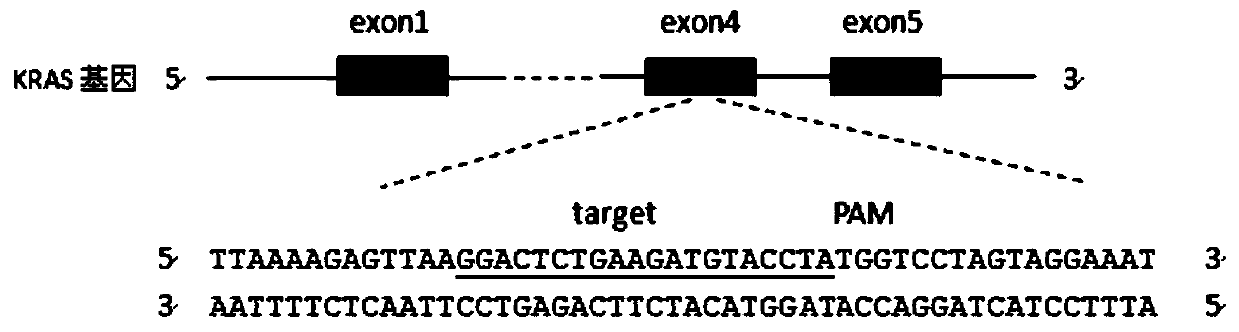
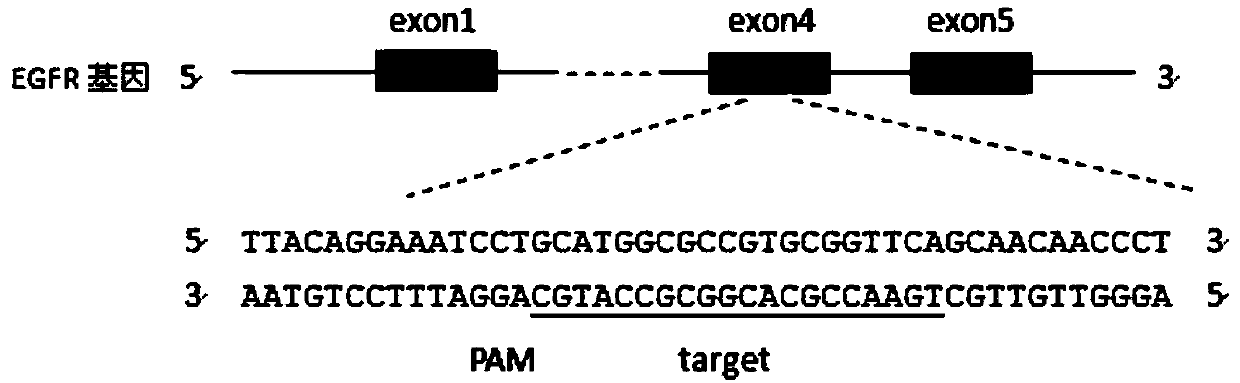
[0035]According to the human KRAS gene sequence (Sequence ID: NM_033360.3) and EGFR gene sequence (Sequence ID: NM_201282.1) given in GeneBank, 10 sgRNAs were designed respectively (the corresponding DNA sequences are shown in SEQ ID NO.1-10 respectively) and shown in SEQ ID NO.11-20). The sgRNAs of the two genes are all targeted at their exon regions, and blasted in UCSC or NCBI to ensure the uniqueness of their target sequences. Of the 10 sgRNAs designed separately, only 2 can effectively knock out KRAS and EGFR, and the first kras-sg1 (corresponding DNA sequence shown in SEQ ID NO.1) and egfr-sg1 (corresponding DNA sequence Shown in SEQ ID NO.11) to explain in detail, such as figure 1 and figure 2 shown.
[0036] 2. Construction of sgRNA oligonucleotide double strands
[0037] Add CACC to the 5' end of the upstream primer of the determined KRAS target sequence, and add AAAC to the 5' end of the downstream primer, so that the double-stranded DN...
Embodiment 2
[0063] T7EN1 digestion experiment:
[0064] The cells collected in Example 1 were lysed, the genomic DNA was extracted with a kit, and finally dissolved in 50 μL of deionized water.
[0065] According to the KRAS gene sequence and EGFR gene sequence published by GenBank, specific primers were designed with Primer 5.0 software, as shown in Table 2:
[0066] Table 2
[0067]
[0068] Use the kit to extract part of the genomic DNA of the cells, and use the extracted genomic DNA of the cells as a template to amplify the target fragment using the specific primers designed in Table 2. Amplification system: 10 μL of 2×PCR Mix, 1 μL of genomic DNA, 1 μL of upstream and downstream primers, plus dd H 2 0 to 20 μL. Reaction program: pre-denaturation at 94°C for 5 minutes; denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 35 seconds, a total of 30 cycles; extension at 72°C for 10 minutes; storage at 4°C.
[0069] 100ng of the purified PCR ...
Embodiment 3
[0071] Western blot experiment:
[0072] Take 1x10 lung cancer cells before and after knockout of KRAS and EGFR 6 After the cells were collected, 20 μL of lysate (50 mM HEPES, PH7.0, 1% NP-40, 5 mM EDTA, 450 mM Nacl, 10 mM Na pyrophosphate and 50 mM NaF) was added, and various fresh inhibitors (1 mM Na orthovanadate, 1 mM PMSF, 10 μg / ml Aprotinin, Leupeptin, pepstatin). After sonication at room temperature, add 1% mercaptoethanol, place at 100°C for 5 minutes and boil for denaturation. On SDS-PAGE gel, load 10 μL of sample per well. After electrophoresis, transfer the protein sample to a nitrocellulose membrane. After transfer, the membrane was washed once with TBST, blocked with 5% skimmed milk powder for 1 hour, washed once with TBST, and the diluted primary antibody was hybridized with the membrane for 2 hours at room temperature or overnight at 4°C. After washing three times with TTBS, the diluted secondary antibody was hybridized with the membrane at room temperature f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com