SgRNA for targeted knockout of RPSA gene and construction method of RPSA gene knockout cell line
A cell line and gene technology, applied in the field of genetic engineering, can solve problems such as lame production performance, low lethality, and high pathogenicity, and achieve the effects of accurate targeting, increasing antigen expression, and promoting replication
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1 Design of sgRNA targeting RPSA gene
[0073] Use the NCBI database to query the RPSA gene sequence, and find the whole genome of the golden hamster (GenBank accession number: NW_004801729), locate the first exon segment of RPSA in the overlapping region of different transcripts in the genome, and use it for target design.
[0074] According to the CRISPR / Cas9 design principle, log in to the CRISPR online design website http: / / crispr-era.stanford.edu / index.jsp to design sgRNA, select 4 pairs of 20bp sgRNA fragments according to the score, and name them respectively: RPSA-sg RNAsp1, RPSA-sgRNAsp2, RPSA-sgRNAsp3, RPSA-sgRNAsp4: add CACC sticky end to the 5' end of the forward sequence of the sgRNA fragment, and add AAAC sticky end to the 5' end of the reverse sequence, as sgRNA oligos targeting RPSA gene polynucleotide (sgRNA-oligo). The sgRNA-oligo was synthesized by Jinweizhi Biotechnology Co., Ltd., and the detailed sequence is shown in Table 1.
[0075] Table...
Embodiment 2
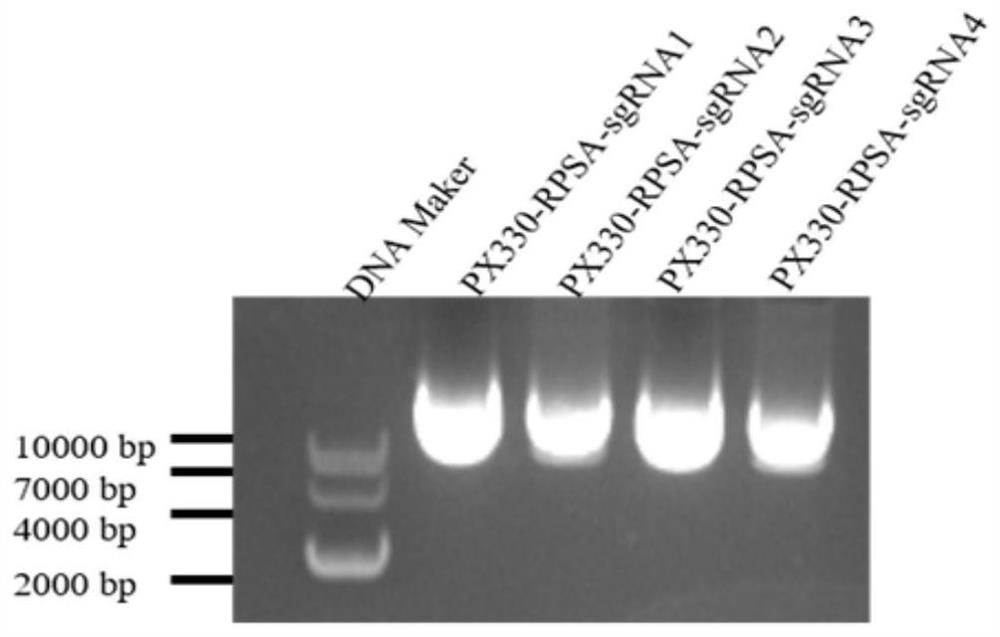
[0078] Example 2 Construction of sgRNA recombinant plasmid PX330-sgRNA
[0079] Obtaining double-stranded sgRNA-oligo: dilute the sgRNA-oligo synthesized in Example 1 to 100 μmol / L, and prepare a total of 10 μl reaction system: upstream primer 2.5, μl; downstream primer, 2.5 μl; ddH 2 O, 4 μl; 10×TaqBuffer, 1 μl. Reaction program: Annealing treatment according to 0.3℃ / s gradient cooling, 95℃, 3min; 95℃, 1min; 85℃, 1min; 75℃, 1min; 65℃, 1min; 55℃, 1min; 45℃, 1min; 35℃ ℃, 1min; 25℃, 1min; 16℃, 1h; anneal the upstream and downstream primers to form a double-stranded sgRNA-oligo.
[0080] Enzyme digestion of PX330 vector plasmid: digest PX330 vector with BBSI restriction endonuclease, and prepare 20 μl of enzyme digestion system as follows: PX330 vector, 5 μl; BBSI, 1 μl; 10×Buffer, 2 μl; ddH 2 O, 12 μl. Place at 37°C for 2h for enzyme digestion. Afterwards, nucleic acid electrophoresis was carried out, and the DNA purification and recovery kit of Promega was used to purify an...
Embodiment 3
[0083] Example 3 Cell Transfection
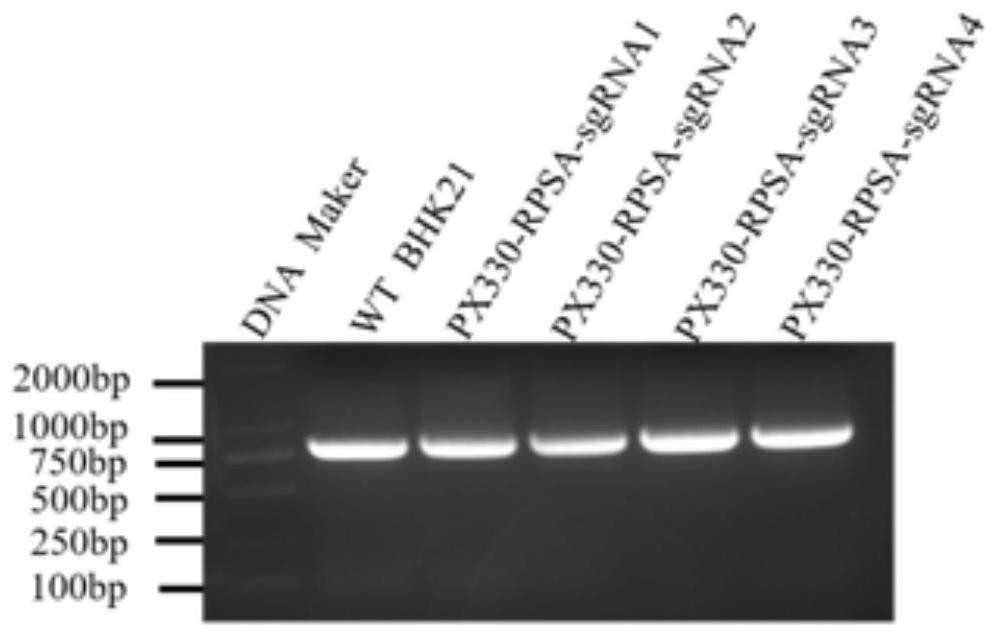
[0084] Resuscitate BHK21 cells in T25 cell flasks before transfection, and culture them in DMEM medium containing 10% FBS and 1% double antibody. In the cell six-well plate, when the degree of cell fusion reached 70% to 80%, the recombinant plasmids successfully constructed in Example 2 (PX330-RPSA-sgRNA1, PX330-RPSA-sgRNA2, PX330-RPSA-sgRNA3, PX330- RPSA-sgRNA4) 2 μg and Lipofectamine3000, 4 μl (according to the ratio of 1 μg: 2 μL) were added to 50 μl of Opti-MEM, and the two were mixed after standing still for 5 minutes. The four liposome-plasmid DNA mixtures were allowed to stand for 15 minutes and then directly added to the cell culture medium. Return the cells to 37°C, 5% CO 2 Cultivate in the incubator for 48h.
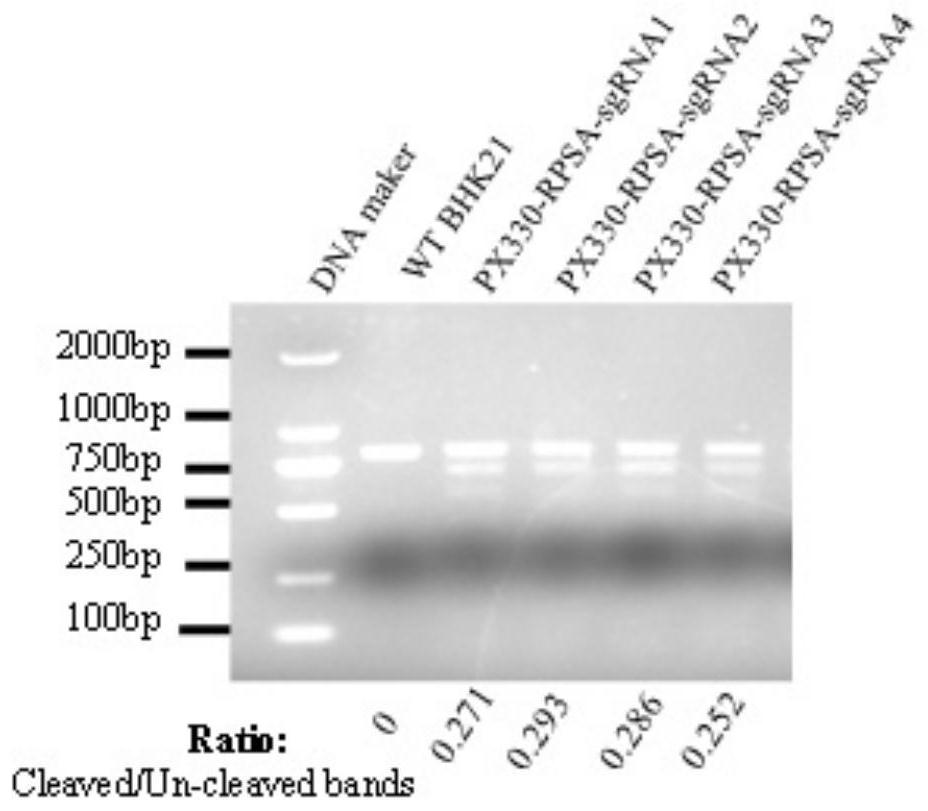
[0085] 1. Cell DNA extraction and SURVEYOR experiment:
[0086] Extract the total DNA of BHK21 cells 48 hours after transfection with the PX330-sgRNA recombinant plasmid according to the operation instructions of the mic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com