A storage and index processing method for genome mutation data
A technology for mutation data and processing methods, which is applied in database indexing, digital data processing, special data processing applications, etc., and can solve the problems of inefficient indexing, increased storage resources, and difficulty in screening.
Active Publication Date: 2021-11-30
HUNAN YEARTH BIOTECHNOLOGICAL CO LTD
View PDF0 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0006] 1) vcf / bcf files can only store a small number of samples in the form of files, and the indexing method based on file pointers is not efficient. In addition to indexing by genomic region, it is difficult to index by mutation site ID, gene name, etc. to index
[0007] 2) When the sample size reaches a certain level, it is difficult to calculate the mutation frequency of a specified site in a large number of samples in real time, and it is difficult to screen out a set of mutations associated with a specified phenotype according to a certain phenotype, or Search for previously recorded pathogenic loci based on a disease name
[0008] 3) When the sample size increases, a large number of vcf / bcf files need to be used for storage, and the occupied storage resources also gradually increase
If you want to mine more data, parsing the vcf file will also become quite complicated
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment 1
[0039] The processing steps of this embodiment are as follows:
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
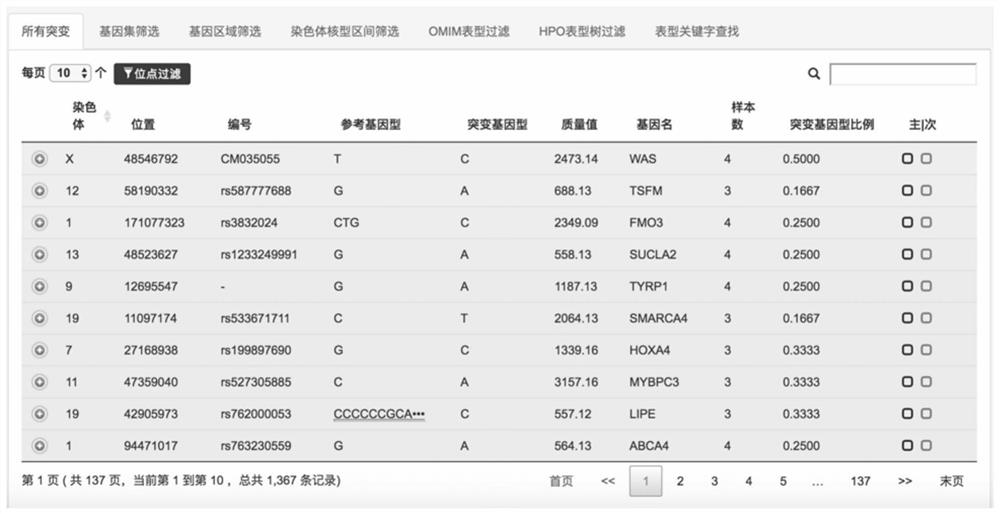
The invention belongs to the technical field of gene data analysis, and in particular relates to a storage and index processing method for genome mutation data. The present invention provides a variety of fast retrieval methods such as retrieving a certain mutation according to mutation site information, retrieving its related mutation list according to single or multiple gene names, retrieving its related mutation list according to single or multiple genome regions, etc. In the field of disease-assisted diagnosis, it is very convenient to quickly find pathogenic sites, and in the field of tumor targeted drug guidance, it is very convenient to quickly find targeted drugs related to tumor mutations, which significantly reduces the processing time of genomic data analysis and interpretation, and greatly reduces The analysis of genomic mutation data is difficult.
Description
technical field [0001] The invention belongs to the technical field of gene data analysis, and in particular relates to a storage and index processing method for genome mutation data. Background technique [0002] In the prior art, genome mutation data is generally stored locally in vcf / bcf file format, and the index of vcf / bcf file is based on the binning indexing algorithm and file pointer to index genome mutation data by region, and only supports vcf / bcf output format . [0003] There are many data storage and indexing technologies, but at present, the storage of genomic mutations is not used in mainstream SQL or NoSQL databases. Usually mutation screening is realized by commonly used vcftools, vcflib, bcftools or self-developed scripts, the functions are relatively single, and personalized screening and customization are difficult. [0004] Although GA4GH defines mutation data to obtain mutation data through RESTful API, it does not define the specific storage format a...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Patents(China)
IPC IPC(8): G06F16/22G06F16/245G16B20/30G16B50/30
CPCG06F16/2255G06F16/245G16B20/30G16B50/30
Inventor 许雄禹黎张刘牛徐根明赵谦
Owner HUNAN YEARTH BIOTECHNOLOGICAL CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com