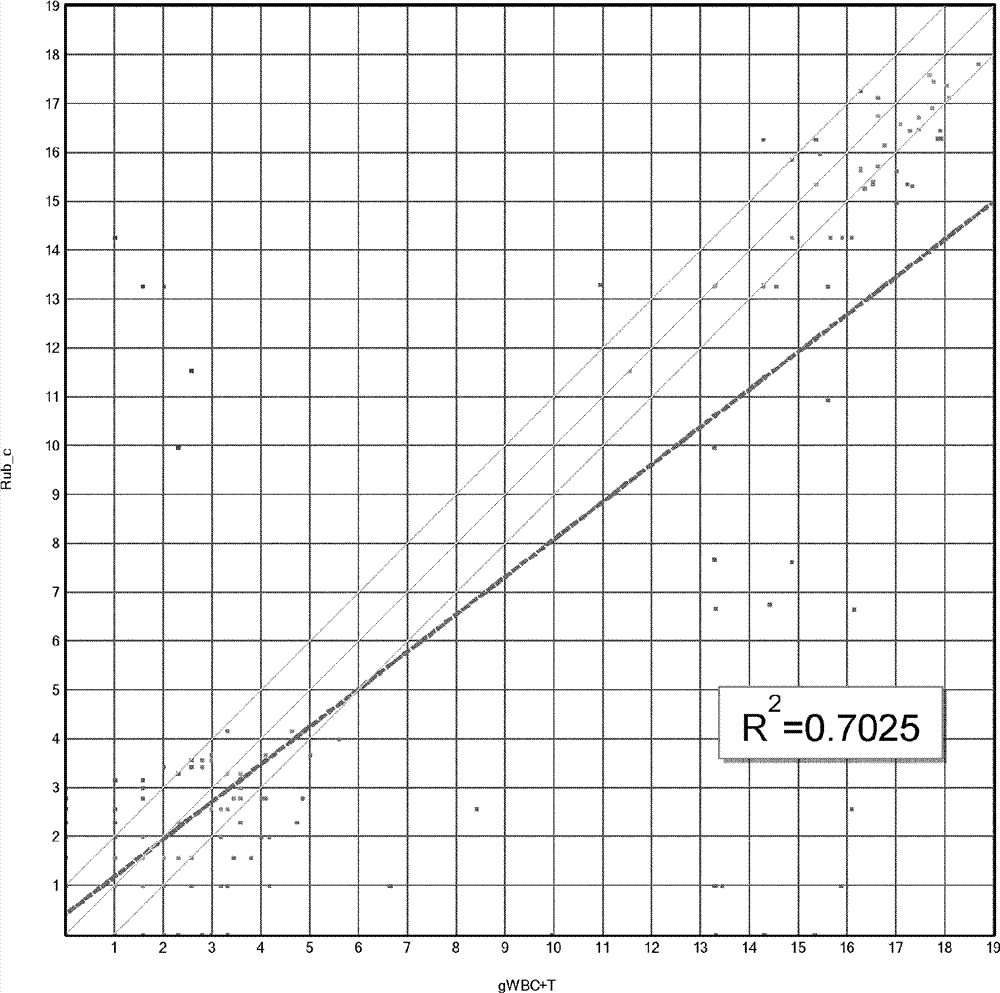
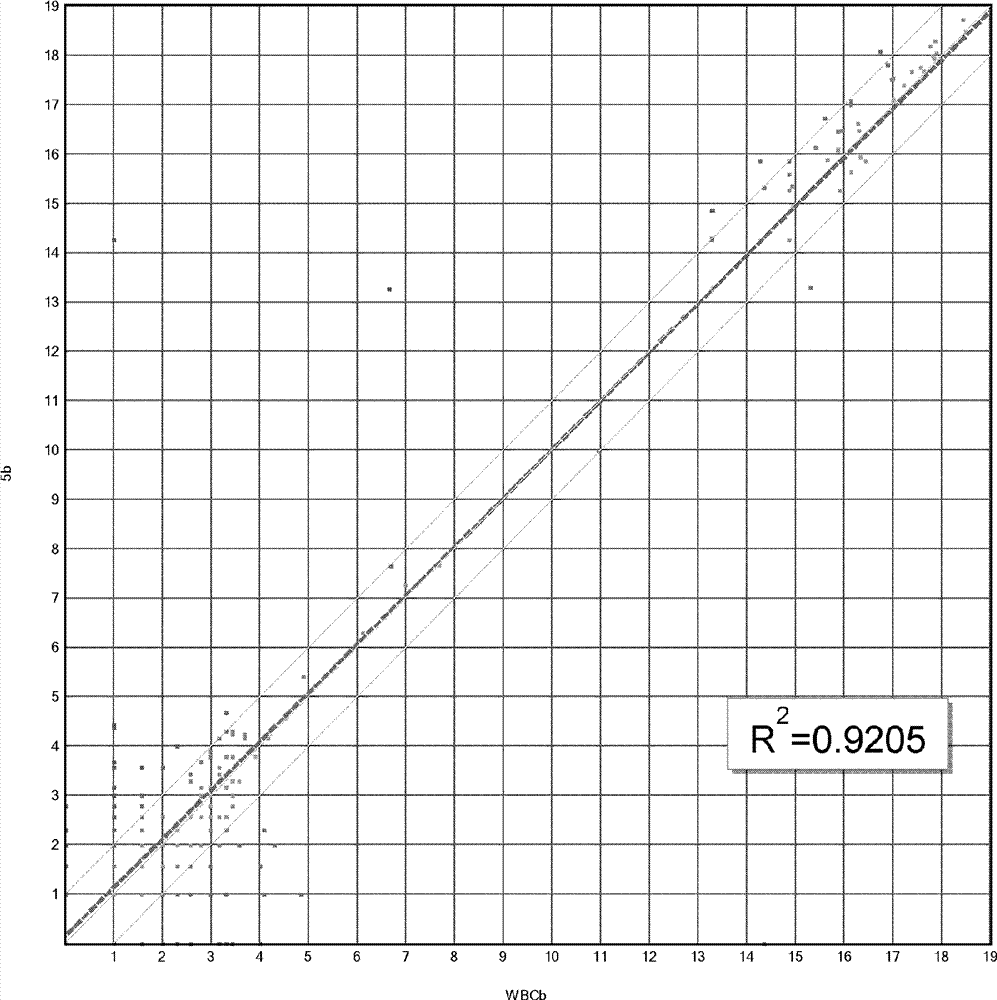
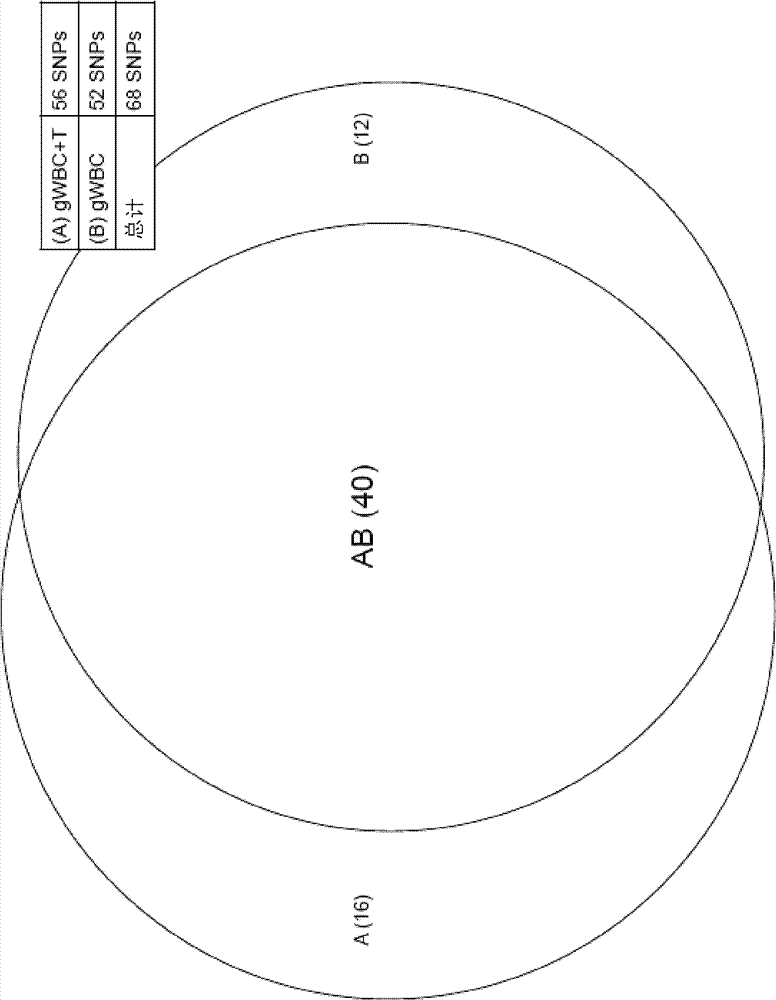
Dual enzymatic amplification
A technology of polymerase and genome, applied in the direction of transferase, biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as wrongly introduced samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0135] Rare cell analysis without whole-genome amplification by massively parallel sequencing
[0136] Materials and methods
[0137] DNA / cell template construction For amplified genomic experiments, pool purified genomic DNA prior to the amplification reaction. For each WGA reaction, 2.4 ng of genomic DNA (~400 cell equivalent based on ~6 pg / cell) was used [5].
[0138] For direct sequencing libraries (DSL), cell pellets are processed to release DNA, which is then used directly in the library construction process without further purification. For DSL experiments, approximately 400 cells were utilized for all reactions. This number is not arbitrary but is based on the average performance of the Cynvenio Biosystems CTC isolation platform (see, e.g., U.S. Patent Publication Nos. 2011 / 0137018; 2011 / 0127222; 2011 / 0003303; 2010 / 0317093; and 2009 / 0053799, for all purposes hereby incorporated by reference in its entirety). The purity of recovered CTCs depends in part on pati...
Embodiment 2
[0210] Dual Enzymatic Amplification to Validate Genomic Mutations in Rare Cell Populations
[0211] In order to measure mutations in the DNA genome of CTCs isolated from 2 to 4 ml whole blood by any technique, sufficient quantity and quality of DNA is important. Typically, recovery of 2 to 10 CTCs from 2 to 4 ml of whole blood can be expected. This number of cells must be processed with excellent recovery to ensure that chromosomes carrying mutations are not lost during processing. Therefore, special methods are required in order to isolate DNA of sufficient quality and quantity. Conventional methods are not useful because they alter DNA genome representation, produce poor quality DNA and / or result in quantities from such rare samples that are unsuitable for use in various molecular assays (such as, but not limited to, QPCR and DNA sequencing) insufficient.
[0212] Isolation of DNA from rare cell populations (eg, small numbers of blood-derived CTC cells) introduces several...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com