A Tamarix salt stress-responsive gene tcsbp1 and its miRNA resistance target rtcsbp1 and its application
A technology of salt stress and genetics, applied in the fields of application, genetic engineering, plant genetic improvement, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Cloning of TcSBP1 gene by RACE technology
[0025] Based on the TcSBP1 transcript of Tamarix RNA-seq data, 5' and 3' RACE primers were designed, and two specific products were amplified by nested PCR, cloned and sequenced by T-vector, and the sequencing results were spliced by overlapping regions, and the complete cDNA was obtained. long. details as follows:
[0026] I. Primer Design
[0027] 3' RACE forward primer (F):
[0028] Outer Primer: 5′-GAATTATTATGTGGCACGAAGGGTAG-3′;
[0029] Inner Primer: 5'-ATTTAACCAGCAATGAACCGCAACA-3';
[0030] 3' RACE reverse primer (R):
[0031] Outer Primer:
[0032] 5'-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3';
[0033]Inner Primer: 5'-CTAATACGACTCACTATAGGGC-3';
[0034] 5'RACE forward primer (F):
[0035] Outer Primer:
[0036] 5'-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3';
[0037] Inner Primer: 5'-CTAATACGACTCACTATAGGGC-3';
[0038] 5'RACE reverse primer (R):
[0039] Outer Primer: 5′-AGCAGCTATGA...
Embodiment 2
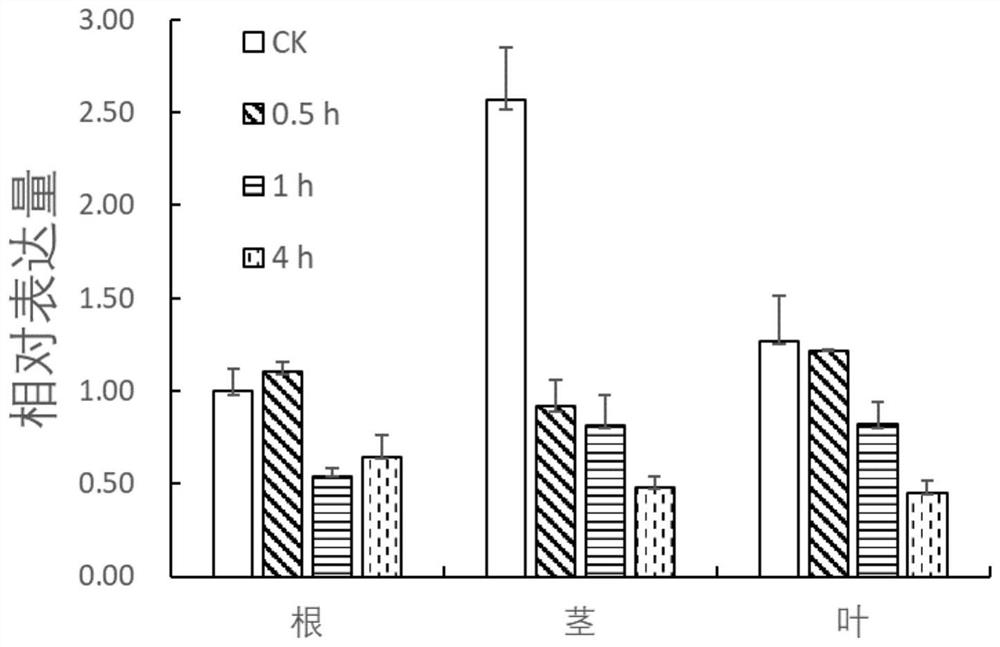
[0075] Example 2: Analysis of TcSBP1 Gene Expression Pattern by Fluorescence Quantitative PCR
[0076] The response to salt stress of TcSBP1 gene was verified by real-time quantitative PCR technology. The fluorescent quantitative PCR primers were designed based on the CDS region of the TcSBP1 gene, and the internal reference primers were designed based on the TIF gene of Tamarix. The sequences are as follows:
[0077] TcSBP1 forward primer: 5′-ATTTAACCAGCAATGAACCGCAACA-3′;
[0078] TcSBP1 reverse primer: 5′-ACCTGCAGGCTTACGTGTGT-3′;
[0079] TIF forward primer: 5'-ACCACAGGAGTGTCCACCACA-3';
[0080] TIF reverse primer: 5'-TGATGCTTTGCGTGCCAGTG-3'.
[0081] Use saturated dye evagreen (Biotium company) and fluorescence quantitative PCR instrument Viia7 (ABI company) to detect the fluorescence intensity of the PCR process in real time. For the specific PCR system, refer to the evagreen instructions, and calculate and determine by comparing the number of cycles that reach the fluor...
Embodiment 3
[0083] The mega-primer PCR method was used for multi-point mutation to obtain the ORF after synonymous mutation of the miR156 response element of TcSBP1. The specific steps are as follows:
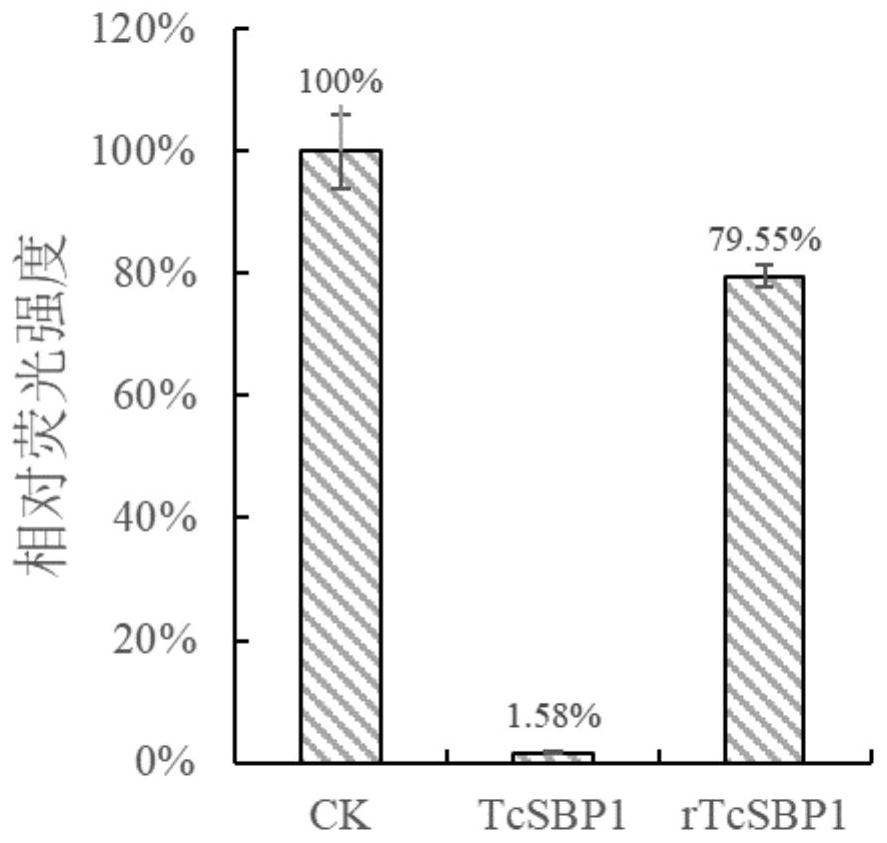
[0084] According to the codon degeneracy, synonymous mutation primers are designed to introduce as many mutation sites as possible, but do not have a poly structure with more than 3 bases, while retaining at least 10nt complementary bases on the flanks. The TcSBP1 mutation primer is 5′- ACAGAATCATCTCATGTAAGCTGTTTCAGTAATCCCATGATC-3', in one round of PCR reaction, a round of PCR product fragments were obtained by amplifying the TcSBP1 mutation primer and the TcSBP1 ORF reverse primer of Example 1. In the second round of PCR reaction, the reaction product of one round was used as the megaprimer, the TcSBP1 ORF forward primer of Example 1, and PCR was performed to obtain the resistance target rTcSBP1 fragment introduced with multiple site mutations. The first and second rounds of PCR were both ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com