A flounder streptococcus dolphin gapdh tandem multi-epitope polypeptide and its application
A Streptococcus iniae, multi-epitope technology, applied in the direction of fusion of polypeptides, peptides, bacteria, etc., can solve the problems of epitope vaccine reports and unsatisfactory effects, and achieve the effect of strong immune protection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: Design of molecular structure of Streptococcus iniae multi-epitope polypeptide
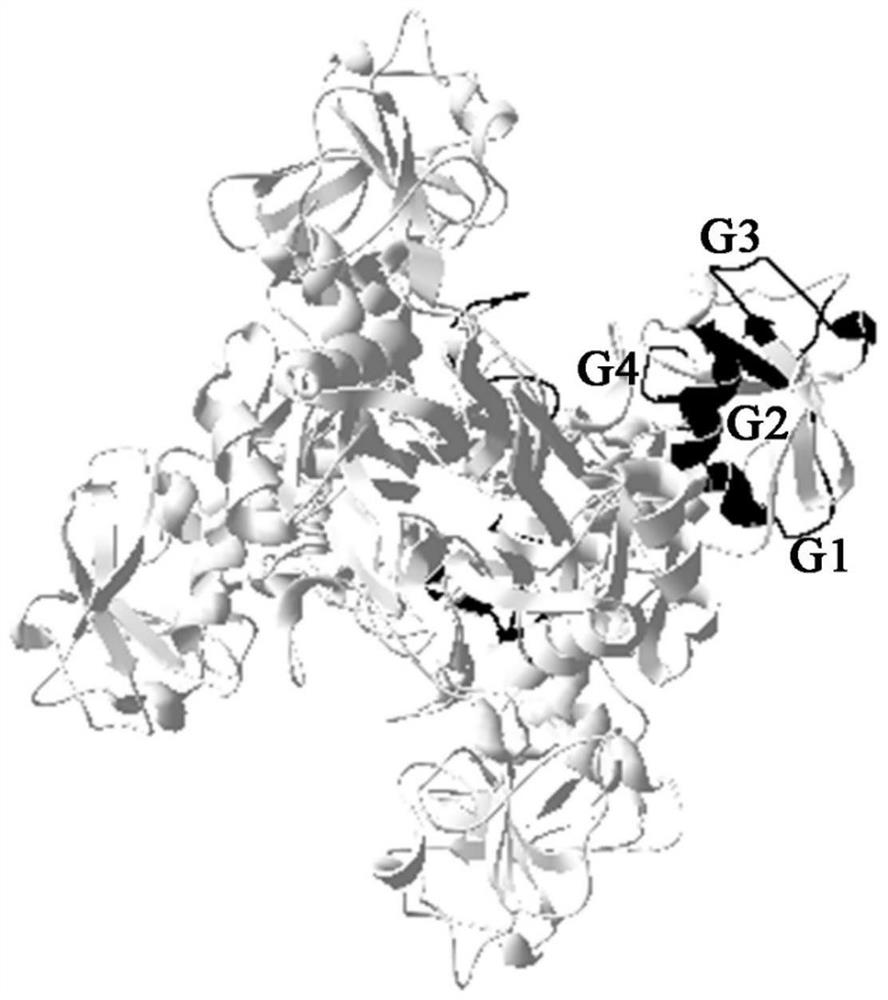
[0032] (1) Using the amino acid sequence of Streptococcus iniae GAPDH protein (ACX85247.1) as the analysis material, the secondary structure of Streptococcus iniae GAPDH protein was predicted by SOPMAServer. The results showed that in the GAPDH gene, random coils accounted for 36.62%, β-turns accounted for 7.32%, α-helixes and extended chains (beta-sheets) accounted for 30.57% and 25.48% respectively. For distribution, see figure 1 .
[0033] (2) Apply DNAStar Protean bioinformatics software and online website (IEDB: http: / / tools.iedb.org / main / bcell / and ProtScale: http: / / us.expasy.org / ) to analyze protein secondary structure The flexible region and plasticity region of the predicted amino acid hydrophilicity (Hydrophilicity Plot -Kyte-Doolittle), flexibility (Flexible Regions-Karplus-Schulz), antigenicity (Antigenic Index-Jameson-Wolf) and surface accessibility (Surface Pr...
Embodiment 2
[0043] Example 2: Construction and induced expression of recombinant expression vector pET-28a-MEPIG
[0044] 1. Construction of recombinant expression vector
[0045] The Streptococcus iniae multi-epitope vaccine molecule designed above is named MEPIG, its amino acid sequence is SEQ ID NO: 8, and the nucleotide sequence corresponding to the linker polypeptide is reversed to the corresponding gene according to the nucleotide tropism of the E. coli expression system sequence, and add BamHI / SacI and SacI / HindIII restriction sites, protective bases, and stop codons to the 5' and 3' ends of the tandem multi-epitope MEPIG gene sequence, respectively, and send them to Shanghai Sangon Bioengineering Co., Ltd. synthesized and cloned it into the pET-28a plasmid to form a recombinant plasmid.
[0046] 2. Induced expression of recombinant multi-epitope polypeptides
[0047] 1) Inoculate the positive bacteria containing the recombinant plasmid after sequencing into 6 mL of LB liquid med...
Embodiment 3
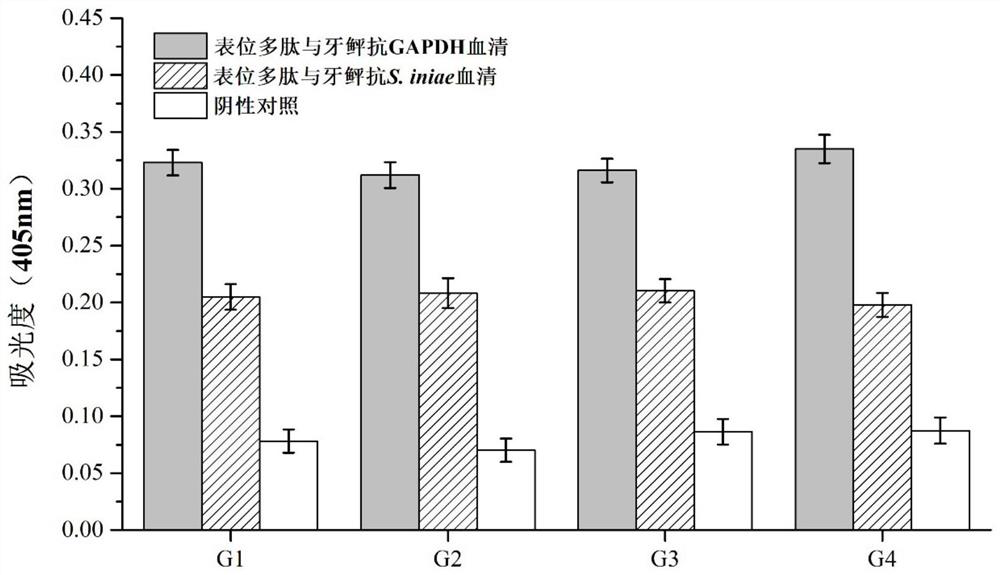
[0049] Example 3: Purification and immunogenicity detection of recombinant multi-epitope polypeptides
[0050] (1) Purification of recombinant protein
[0051] Centrifuge the bacterial solution confirmed to be successfully induced with a centrifuge (8000×g, 10min, 4°C), discard the supernatant, collect the bacterial cells, wash once with PBS, and use about 40-50mL of Binding buffer (Na 2 HPO 4 14.5g; NaH 2 PO 4 1.48g; NaCl 29.3g; urea 480g; 40mM imidazole; pH 7.4, dilute to 1L with ultrapure water, and filter with membrane) to resuspend, place on ice, and crush with ultrasonic breaker (3s on, 3s off, power 37% ) to light yellow clear liquid, after centrifugation at 8000×g for 5min, take the supernatant, and store it at 4°C for later use; connect the sample cup, His-tagged protein affinity chromatography column, etc., and check the airtightness of the device; wash with ultrapure water Instrument pipeline A, B and protein affinity chromatography column, and then use the pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com