Streptococcus iniae PDHA1 multi-epitope polypeptide
A kind of Streptococcus iniae, multi-epitope technology, applied in the direction of antibacterial drugs, bacterial antigen components, enzymes, etc., to achieve the effect of strong immune protection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: Bioinformatics analysis of B cell linear epitopes of PDHA1 protein
[0024] 1. Predicted epitopes
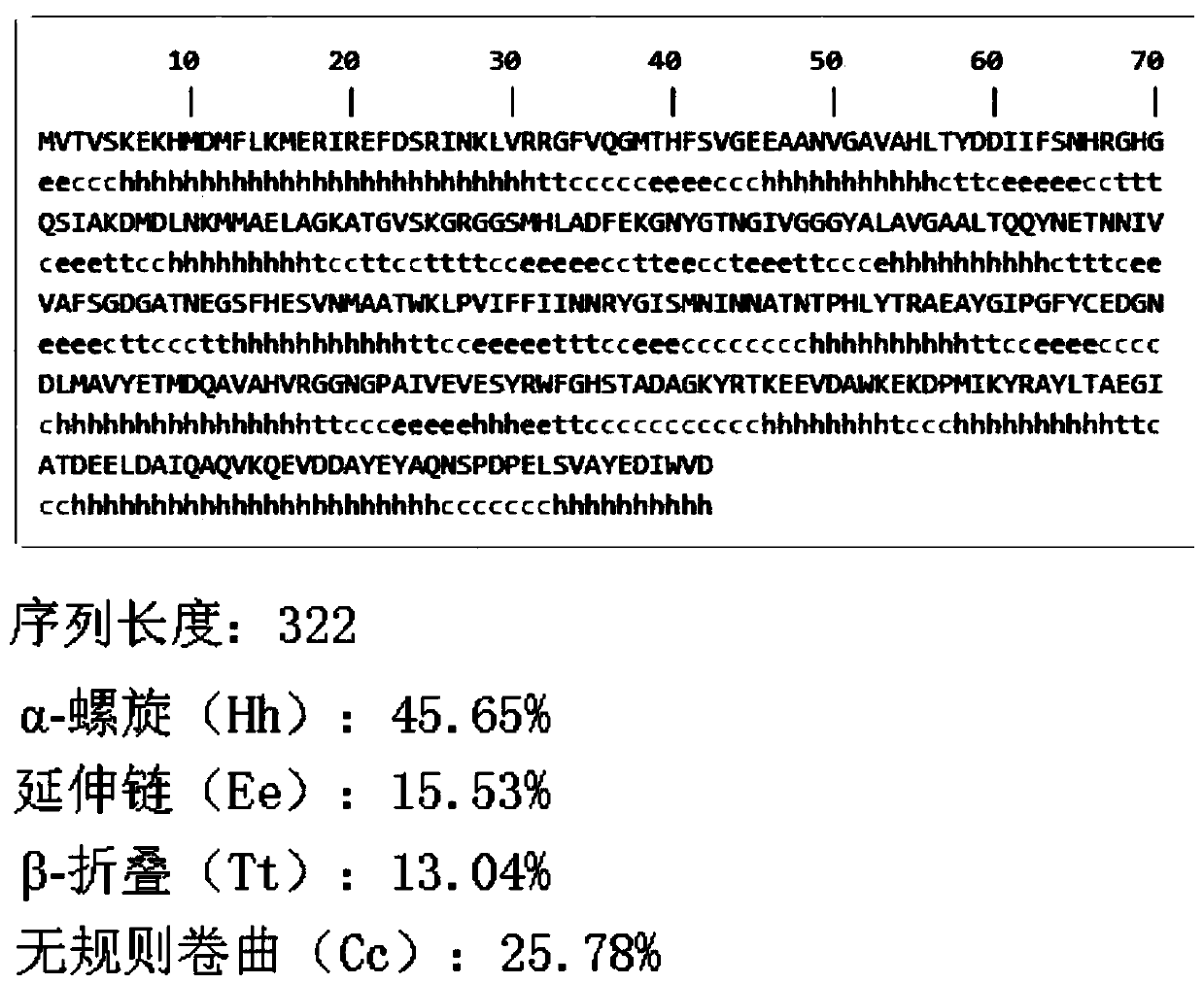
[0025] 1) Using the amino acid sequence of Streptococcus iniae PDHA1 protein (WP_003099247.1) as the analysis material, the secondary structure of Streptococcus iniae PDHA1 protein was predicted using SOPMAServer. The results showed that random coils accounted for 25.78% and β-turns accounted for 13.04% of the PDHA1 gene. The α-helix and extended chain, namely β-sheet, account for 45.65% and 15.53% respectively. For the distribution of various structures in the amino acid sequence of Streptococcus iniae PDHA1 protein, see figure 1 .
[0026] 2) Apply DNAStar Protean bioinformatics software and online website (IEDB: http: / / tools.iedb.org / main / bcell / and ProtScale: http: / / us.expasy.org / ) to analyze protein secondary structure Flexible and plastic regions, predicting amino acid hydrophilicity (Hydrophilicity Plot-Kyte-Doolittle), flexibility (Flexible Regions-Ka...
Embodiment 2
[0046] Example 2: Design of tandem multi-epitope polypeptide molecular structure and construction of recombinant expression vector
[0047] 1. Construction of tandem multi-epitope polypeptides
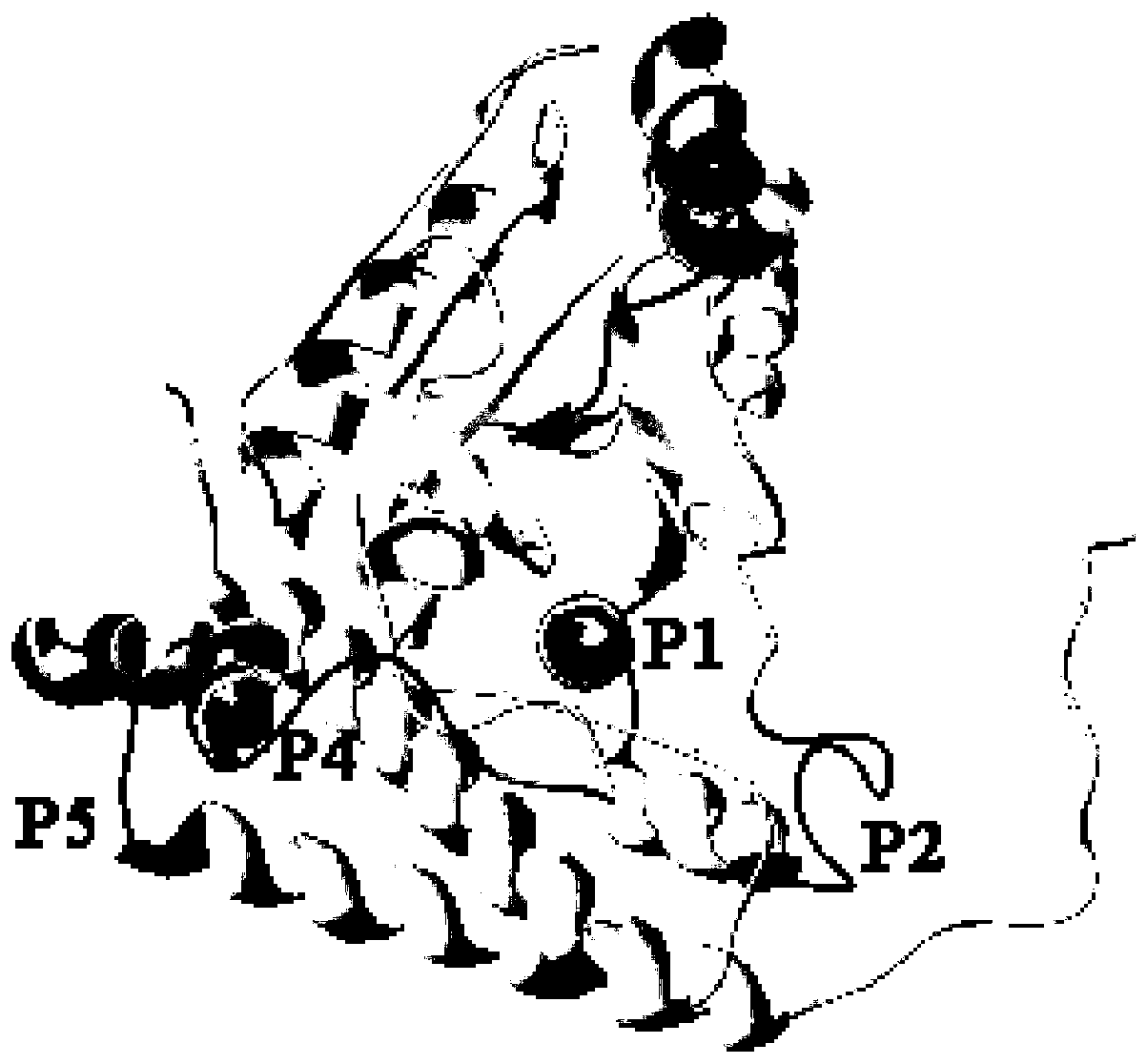
[0048] (1) The identified antigenic epitopes are connected in series using different polypeptide linkers (GGGG, EAAAK, AAY, KK, GPGPG) and different connection methods to determine the best connection method. The results show that after PDHA1 is connected in series with the connection sequence of KK linker (P1-KK-P2-KK-P3-KK-P4), each epitope is relatively independent, and the antigenic parameters are good ( Figure 4 ). The tandem multi-epitope polypeptide is named MEPIP, and its amino acid sequence is SEQ ID NO:5:
[0049] NHRGHGQSIAKDMDKKAGKATGVSKGRGGSKKKYRTKEEVDAWKEKKKRAYLTAEGIATDEE.
[0050] (2) Obtain the gene sequence of each epitope, and reverse the nucleotide sequence corresponding to the linker polypeptide into the corresponding gene sequence (SEQ ID NO: 6) according to th...
Embodiment 3
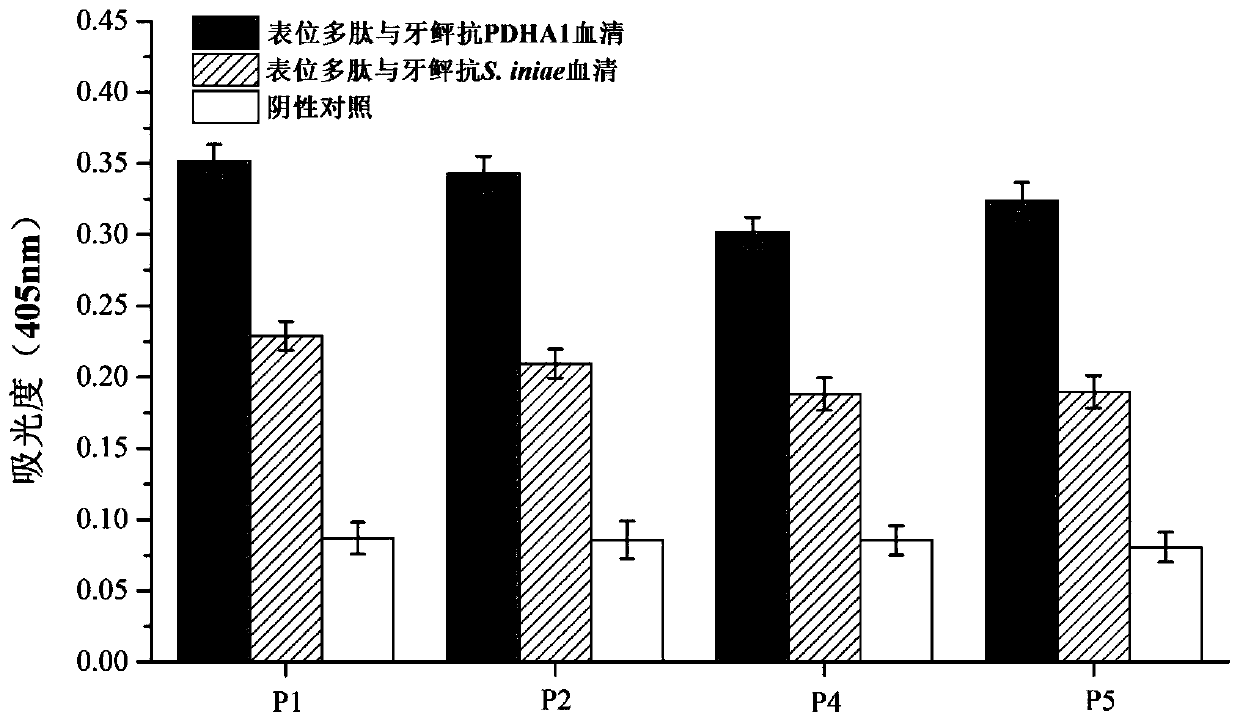
[0073] Example 3: Application of PDHA1 multi-epitope vaccine in providing immune protection for flounder against S.iniae infection
[0074] (1) Immunization and sampling of flounder
[0075] ①Experimental settings The recombinant protein (rMEPIP) of the multi-epitope polypeptide MEPIP prepared in Example 2, the recombinant subunit vaccine (rPDHA1), inactivated Streptococcus iniae (FKC) and the PBS negative control consisted of 4 groups, each group of flounder 100 Finally, the protein concentration was determined by BCA method and the protein concentration was adjusted to 2 mg / mL with sterile PBS, and the vaccine protein and Freund's complete adjuvant were lubricated with a syringe lubricated with Freund's complete adjuvant, respectively, connected with a connecting tube, and mixed with an injection emulsifier until the mixture does not diffuse in water. The negative control group was injected with an equal volume of adjuvant and PBS.
[0076] ② Before immunization and 1d, 3d...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com