A new method for estimating the species composition of aquatic animal genomes based on the admixture-mcp model
A technology of aquatic animals and genome, applied in the fields of genomics, proteomics, bioinformatics, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
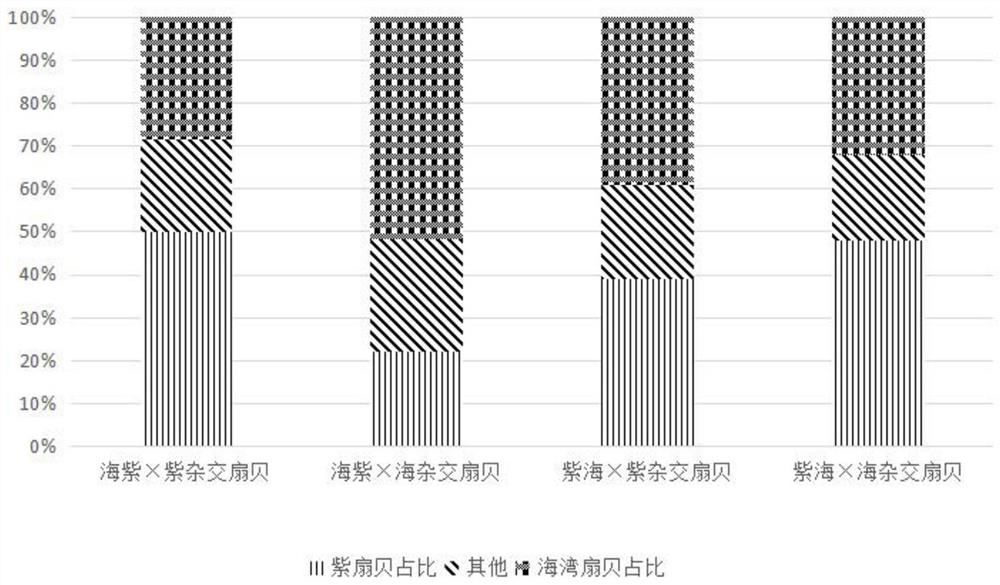
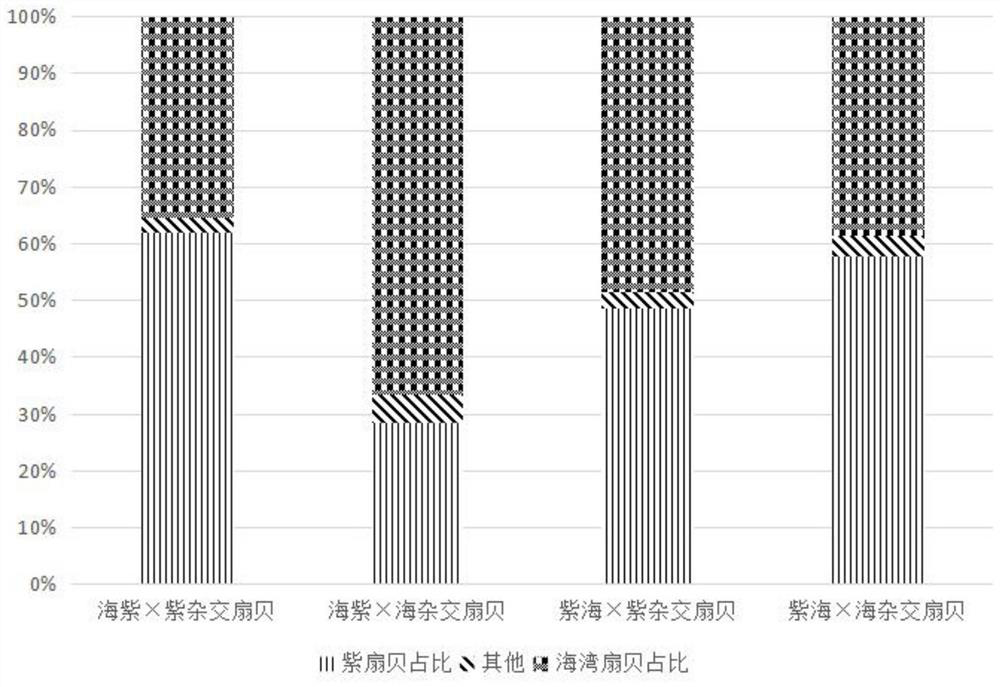
[0052] The specific implementation manners of the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments. The following bivalve scallop among aquatic animals is taken as an example to illustrate the present invention, but not to limit the scope of the present invention.
[0053] Gulf scallop is one of the most important cultured shellfish in my country, but the germplasm degradation problem in recent years has become a serious obstacle restricting the development of my country's bay scallop aquaculture industry. Both purple scallop and bay scallop belong to the excellent species of bay scallop, and they have complementary traits. Through interspecific hybridization, it is expected to breed scallops with fast growth speed, large individual size and wide temperature adaptation range, which can improve the germplasm of bay scallop and improve the quality of scallop. The economic output of aquaculture.
[0054] Th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com