SNP molecular markers related to barley grain β-glucan content and their application
A technology of molecular markers and dextran, which is applied in the fields of molecular biology and genetic breeding, can solve the problems of non-specificity of β-glucan, high price of kits, and low measurement results, so as to improve identification efficiency and accuracy High degree, convenient and stable amplification, accurate and efficient detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] (1) Test materials
[0028] Using 100 global core barley germplasm resources as materials, they were planted and harvested in Hangzhou and Ningbo from 2018 to 2019, and the seeds were dried and stored for later use.
[0029] (2) Determination of traits
[0030] Grind the above-mentioned harvested seeds into powder with a sample grinder, and place them in a desiccator for dry storage for subsequent content determination. The β-glucan determination kit of Megazyme Company is used to determine the grain β-glucan of 100 parts of barley materials content.
[0031] (3) GWAS analysis and SNP molecular marker determination
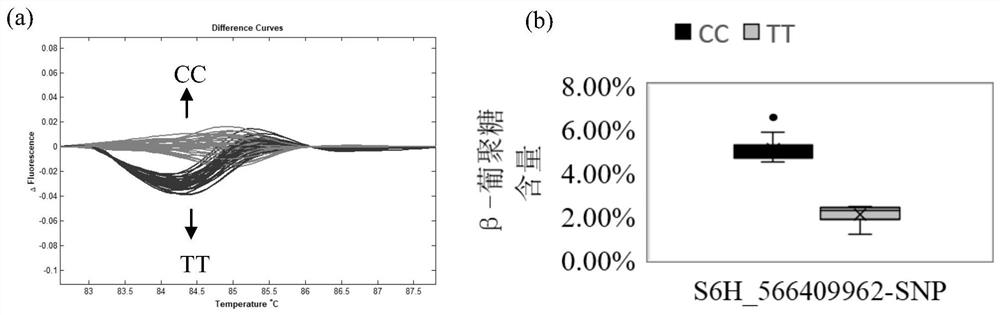
[0032] Combined with the β-glucan content determined above and 279,515 SNP markers in this population, TASSEL software was used for GWAS analysis. The results showed that a SNP marker located on chromosome 6 was significantly associated with barley grain β-glucan content. The SNP is located at base 566409962 of chromosome 6 and can be detected repeatedly...
Embodiment 2
[0034] (1) Test materials
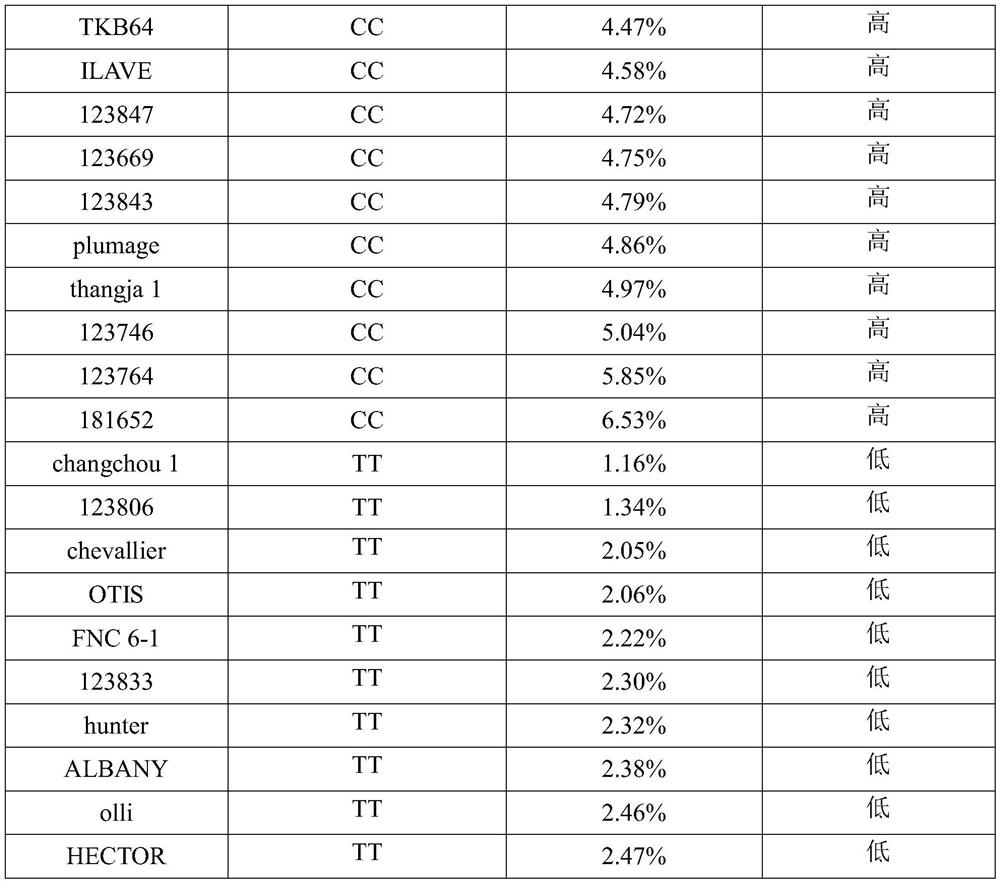
[0035] Using 20 global core germplasm resources as materials, the determination of grain β-glucan content and the analysis of S6H_566409962-SNP target region were carried out respectively. Specifically, as shown in Table 1, it includes 10 parts of high β-glucan material and 10 parts of low β-glucan material.
[0036] Table 1 20 barley germplasm materials with different β-glucan contents
[0037]
[0038]
[0039] (2) Acquisition of SNP markers
[0040] According to the above SNP site information, combined with the barley genome sequence information, SNP marker primers were developed. The upstream primer F is: 5'-CCGACTTCGTCTACGTGCTC-3'; the downstream primer R is: 5'-GGAGTTGCGGTACTTGTCGG-3', and the amplification size is 98bp , the SNP is located at the 47th bp of the amplified fragment.
[0041] Using the above primer pair and the following technique, the variation of the base at 566409962 on chromosome 6 of barley is detected, so as to re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com