Systems and methods for producing retinal progenitors
A technology of retinal progenitor cells and stem cells, applied in the field of systems and methods for generating retinal progenitor cells, capable of solving problems such as not showing effectiveness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Embodiment 1
[0111] Cell line development of HEKs overexpressing LN-523 and LN-323
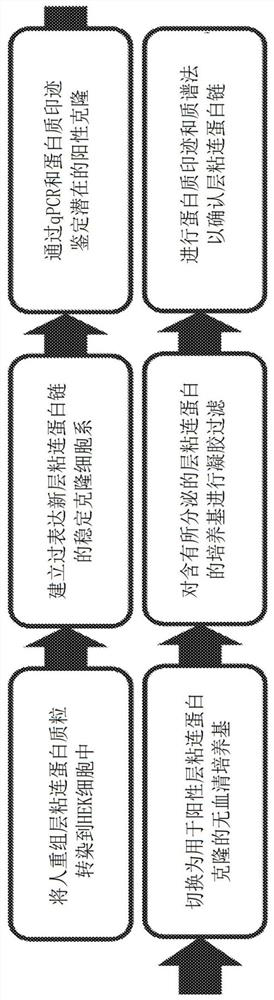
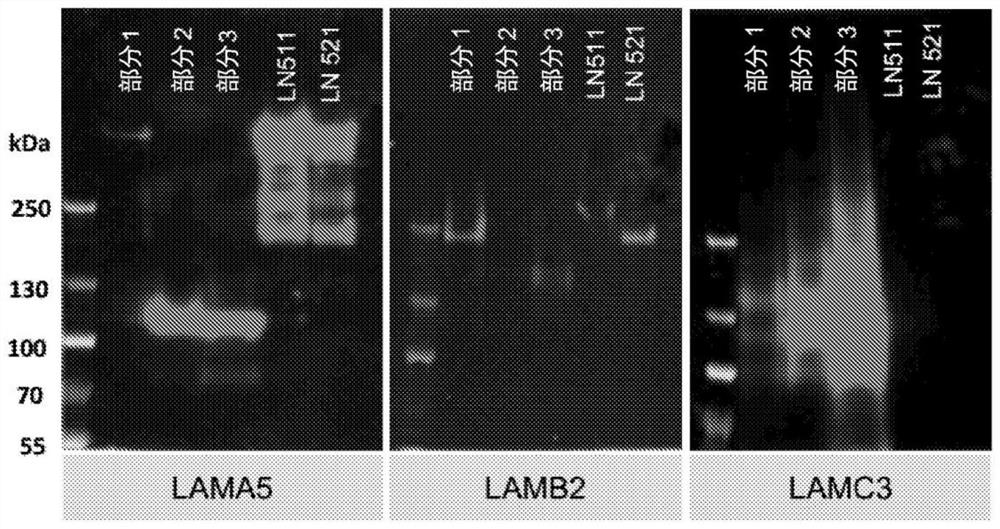
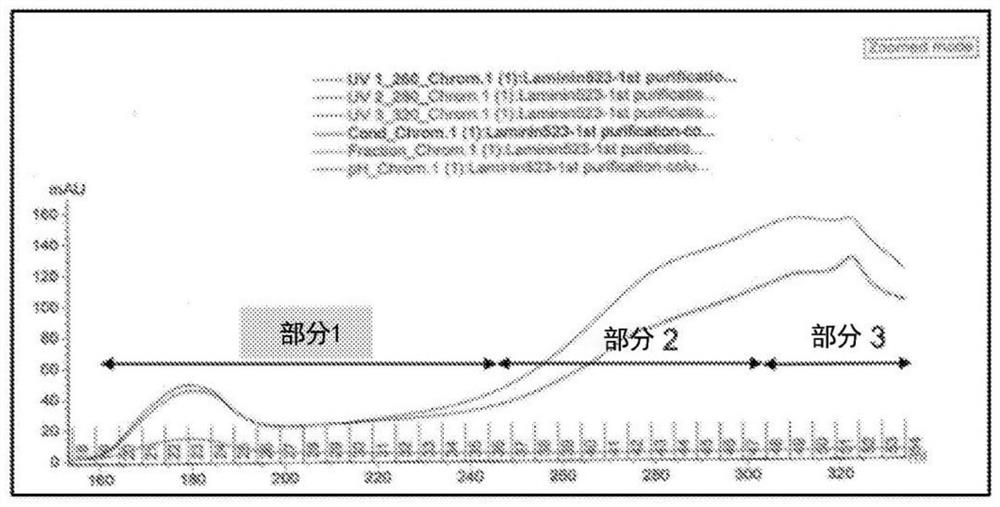
[0112] exist Figure 1A A flowchart describing the steps for producing recombinant laminin-523 and laminin-323 is provided in . Briefly, human recombinant laminin expression plasmids encoding laminin α-3, β-2 and γ-3 chains and containing three different antibiotic resistance genes were generated and sequentially transfected into HEK cells. To generate LN-523, expression plasmids encoding laminin α-5, β-2 and γ-3 chains were sequentially transfected into HEK cells. Stable clonal cell lines overexpressing the desired laminin chains were established. Third, identify potential positive clones using qPCR and Western blotting. Fourth, switch the cell culture medium to serum-free medium for positive laminin clones. Fifth, the cell culture medium containing the secreted laminin is gel filtered. Finally, western blotting and mass spectrometry were performed to confirm the identity of the laminin chains.
[0...
Embodiment 2
[0144] Two independent single-cell RNA-sequencing datasets were generated using day 20 and day 30 retinal progenitor cells derived from H1 embryonic stem cells. H1 cells were cultured with the laminin protocol of Example 1 for 20 and 30 days. In each case, RNA was isolated using a 10× Genomics kit and sequenced using the Illumina Hi-Seq3000 sequencing platform. Reads were mapped to the human genome (Ensembl version 90) and quantified using Cell Ranger 2.1.1 10x Genomics software. A custom reference transcriptome generated by genotype (protein coding, lincRNA, and antisense) filtering of the Ensembl transcriptome was provided to the Cell Ranger software. Cell Ranger was run with the expected cell number parameter (expected cells) set to 3000.
[0145] The estimated cell numbers for the Cell Ranger at day 20 and day 30 were 1,790 and 3,881, respectively. The total reads on days 20 and 30 were 631,803,330 and 687,721,993, respectively. The percentage of reads mapped to the hu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com