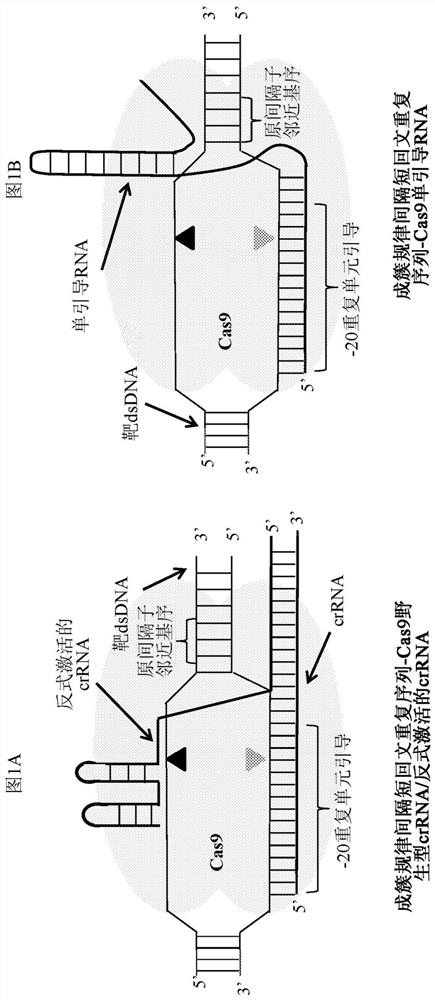
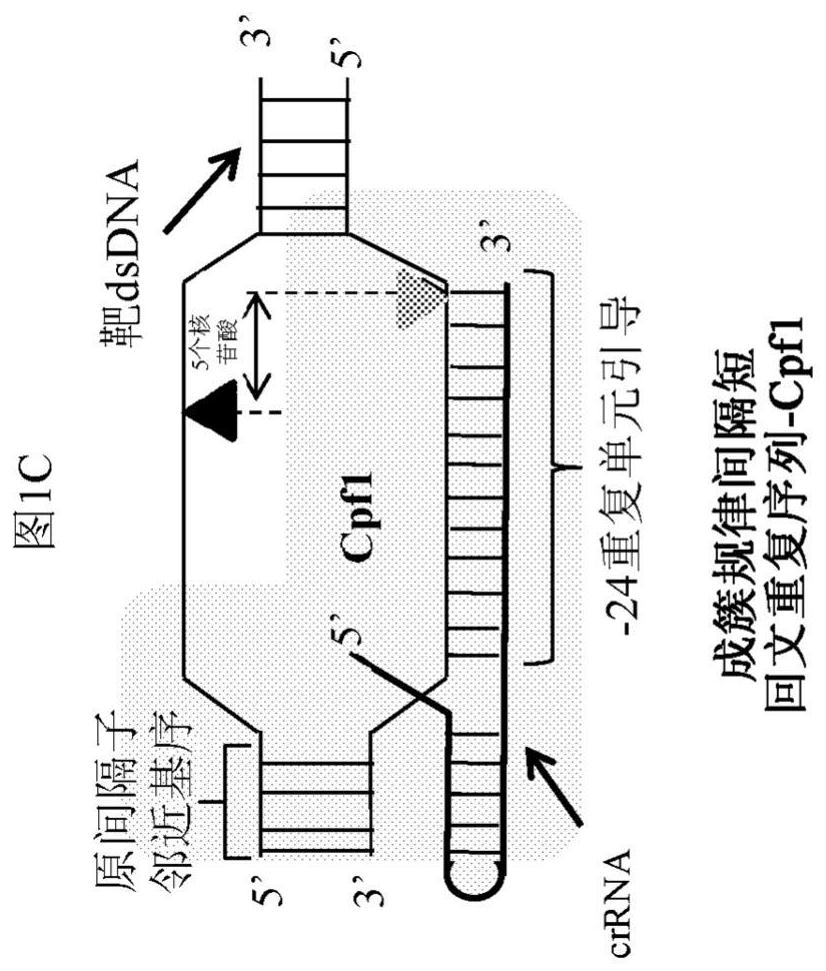
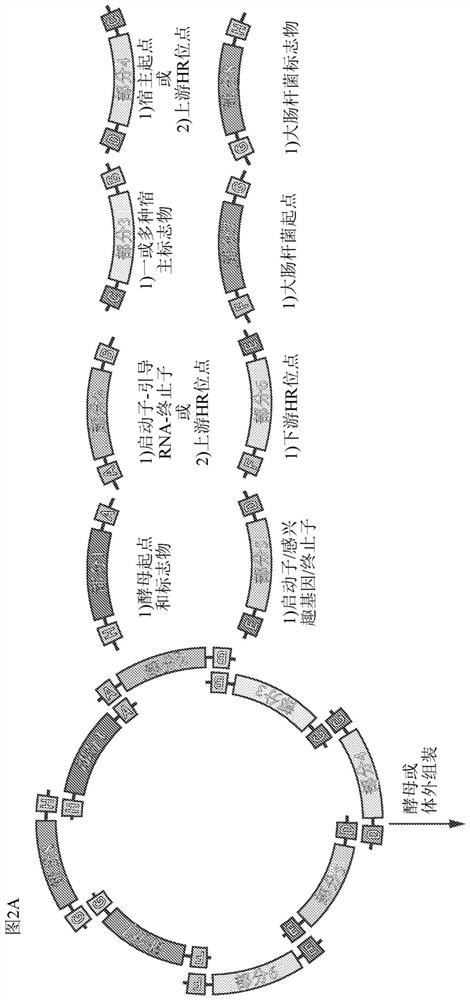
APPLICATIONS OF CRISPRi IN HIGH THROUGHPUT METABOLIC ENGINEERING
A modular and constructive technology, applied in the high-throughput field, can solve problems such as expensive, low-efficiency effects, and off-target mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0380] The following examples are given for the purpose of illustrating various embodiments of the present disclosure and are not meant to limit the present disclosure in any way. As defined by the scope of the claims, example variations and other uses will occur to those skilled in the art that are encompassed within the spirit of the present disclosure.
example 1
[0381] Example 1: One-pot in vitro modular CRISPR cloning
[0382] This example describes the generation of plasmid 13001009086 (SEQ ID NO: 82) by transferring an insert from one plasmid to another in a one-pot reaction. see Figure 4 .
[0383] Both plasmids carry cloning tags (cTAG K[SEQ ID NO:78] / cTAGL[SEQ ID NO:79] and cTAGK'[SEQ ID NO:80] / cTAGL'[SEQ ID NO:80] ID NO:81]). To drive cloning responses to edited plasmids, the Cpf1 spacer was in opposite orientation (K / K' and L / L', respectively) on the acceptor and donor plasmids. This inside-out / outside-in digestion removes the Cpf1 spacer in the final product, thereby eliminating re-cleavage of the desired product (see Figure 4 Curved arrows in , which depict inside-out digestion in the '485 plasmid and outside-in digestion in the '784 plasmid). The Cas9 spacer is retained, enabling iterative editing at this site. Thus, large modular constructs allow for a fast one-pot reaction scheme for iterative editing.
[0384] T...
example 2
[0387] Example 2: In vitro modular CRISPR cloning
[0388] This example is designed to demonstrate the flexibility of CRISPR cloning. As an initial step, several resistance plasmids encoding kanamycin or chloramphenicol resistance genes were generated from source vectors pzHR039 (SEQ ID No: 100) and 13000223370 (SEQ ID No: 101 ), respectively. The kanamycin resistance plasmids were each designed to contain various Cpf1 landing sites flanking the GFP gene (when digested, these plasmids produced a "kanamycin-resistant plasmid backbone"). The chloramphenicol resistance plasmids were each designed to contain various Cpf1 landing sites flanking the chloramphenicol resistance gene (when digested, these plasmids produced "chloramphenicol-resistant inserts"). The sequence and vector map of each plasmid used in this example is disclosed in Table 6.
[0389] Each kanamycin- and chloramphenicol-resistant plasmid was initially linearized with type II restriction enzymes KpnI-HF and PvuI...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com