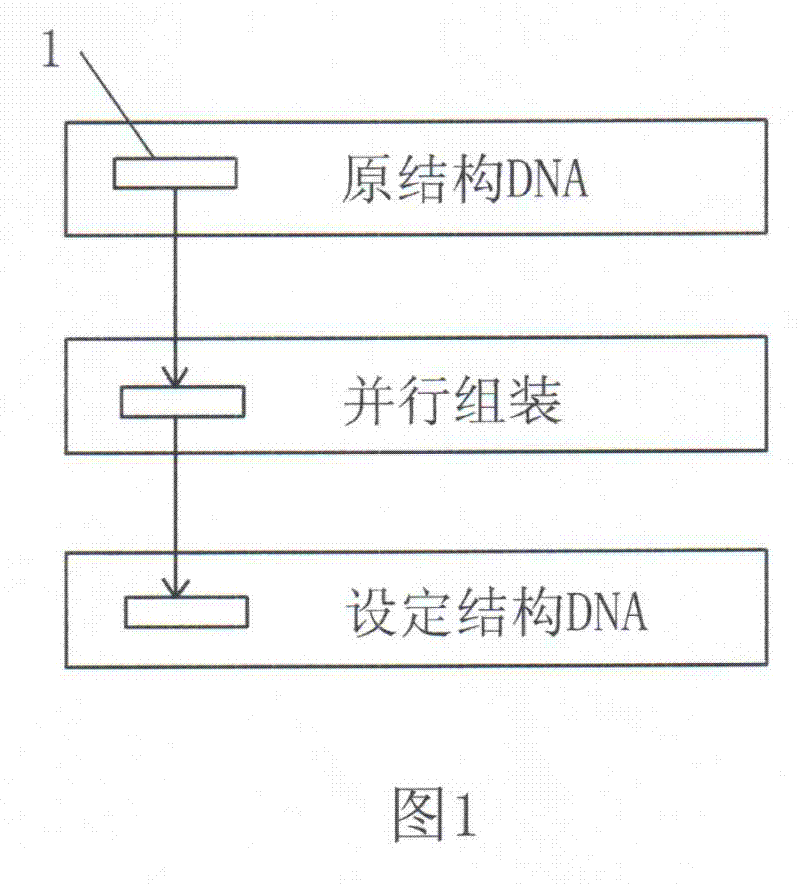
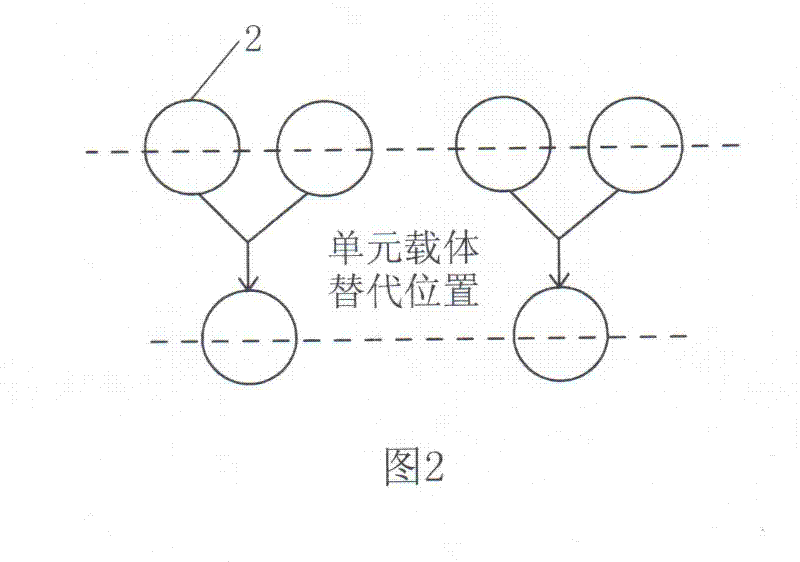
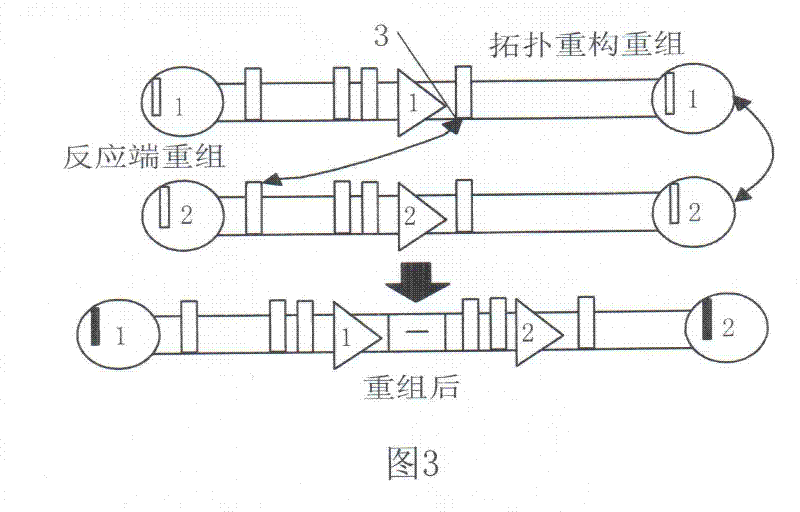
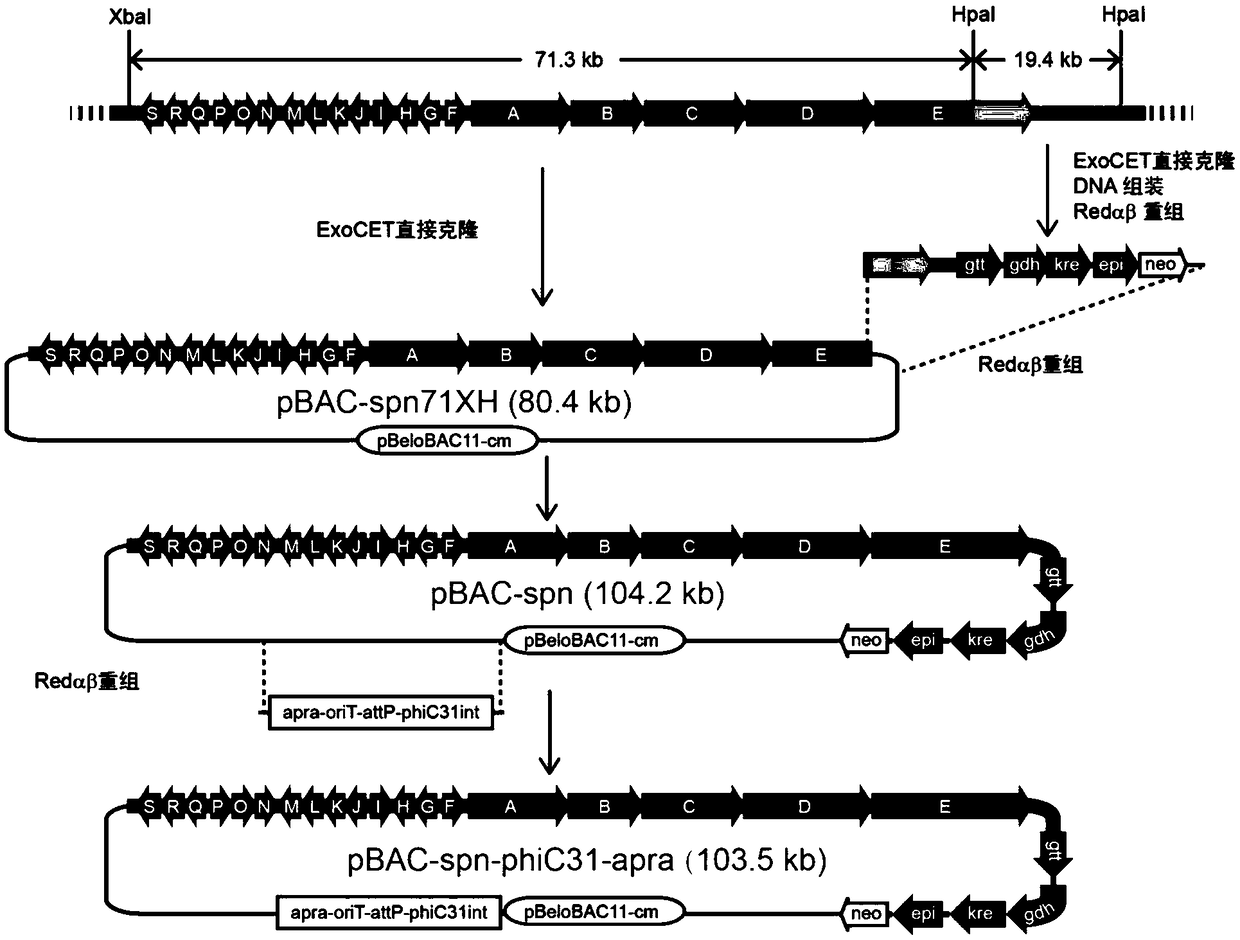
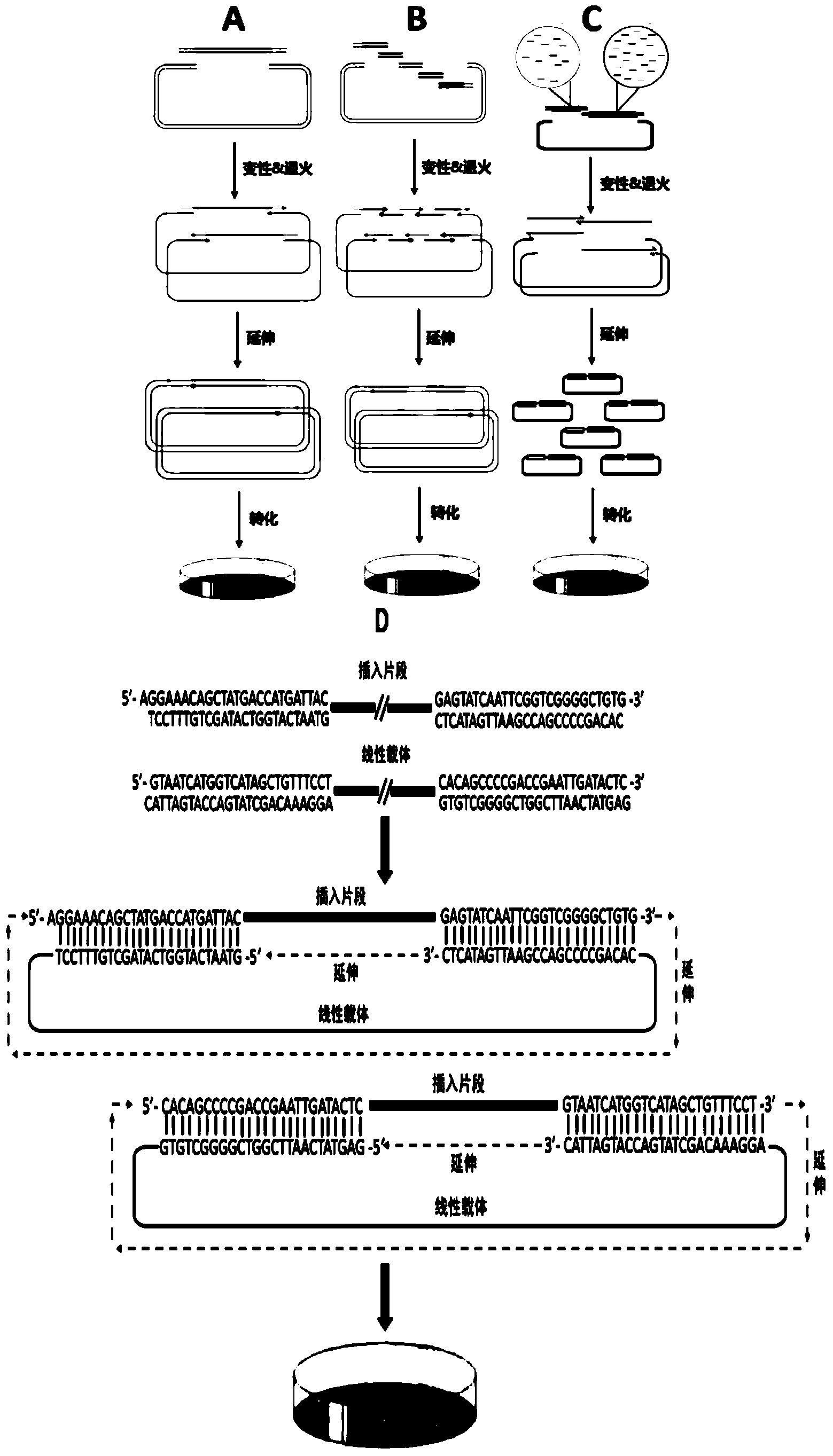
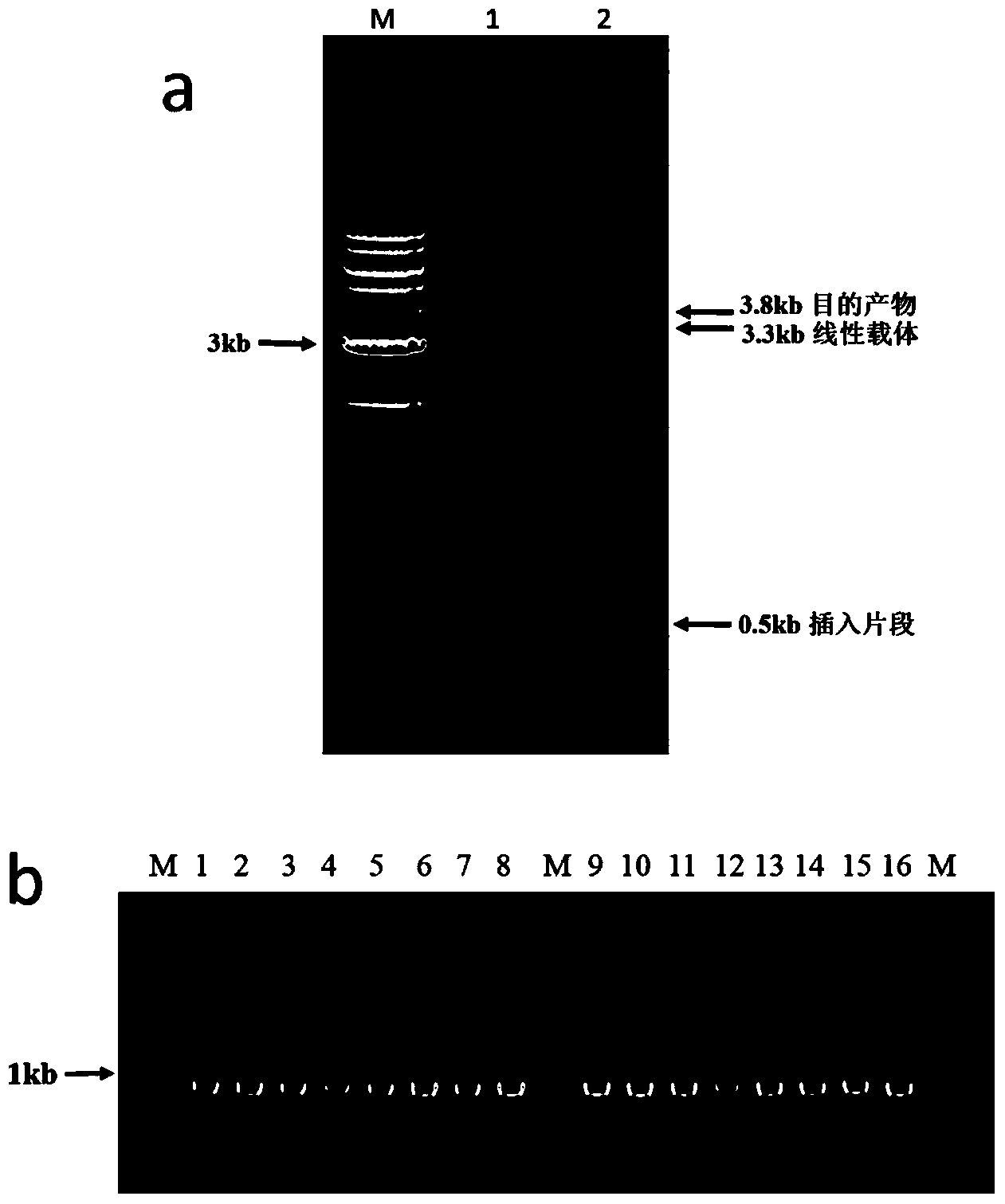
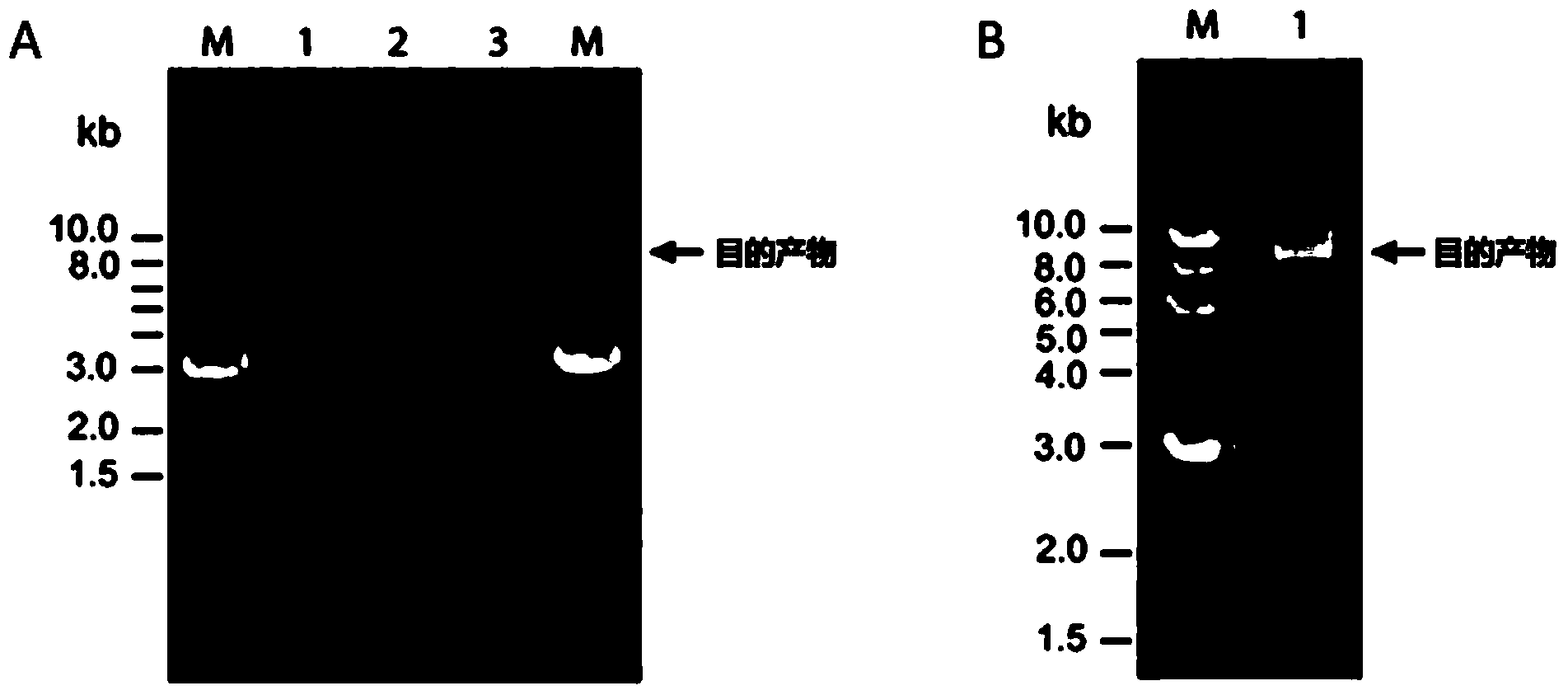
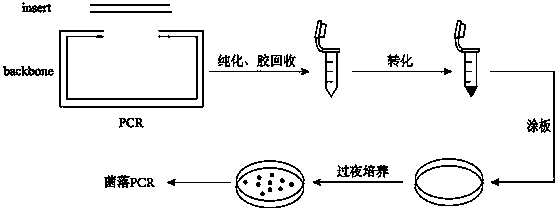
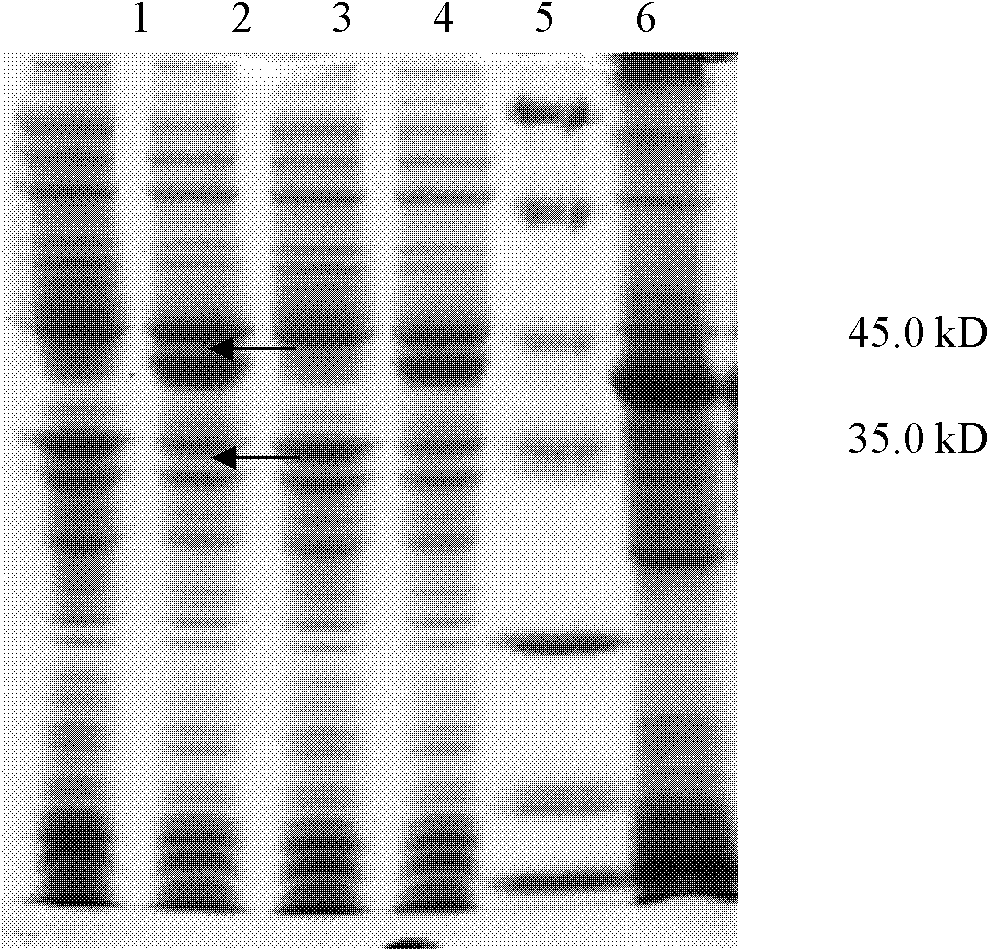Patents
Literature
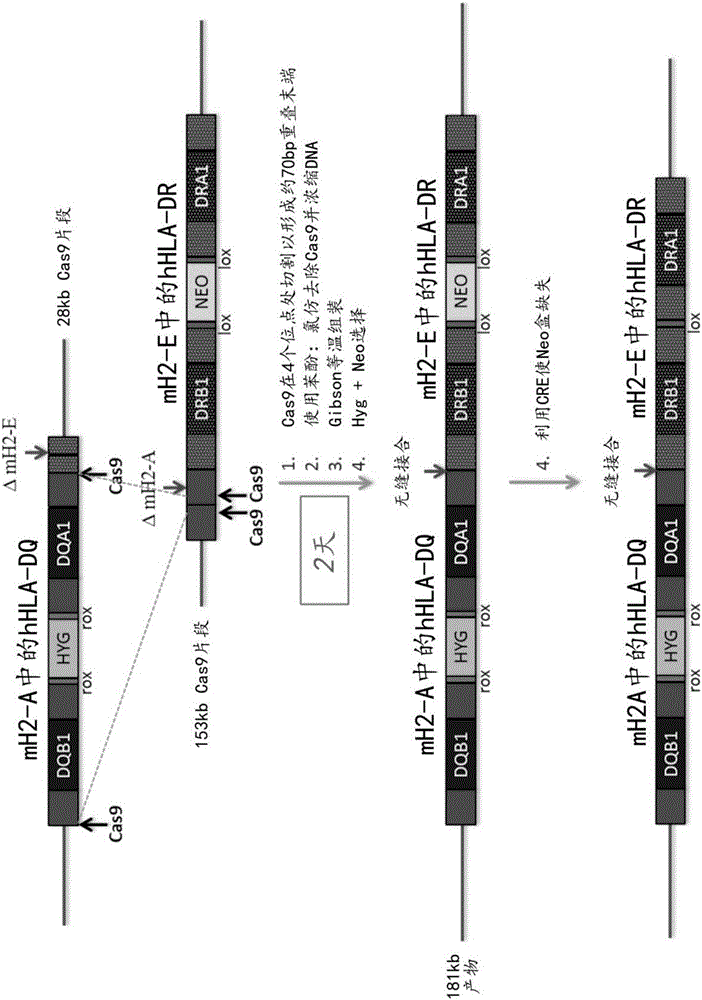
69 results about "Dna assembly" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
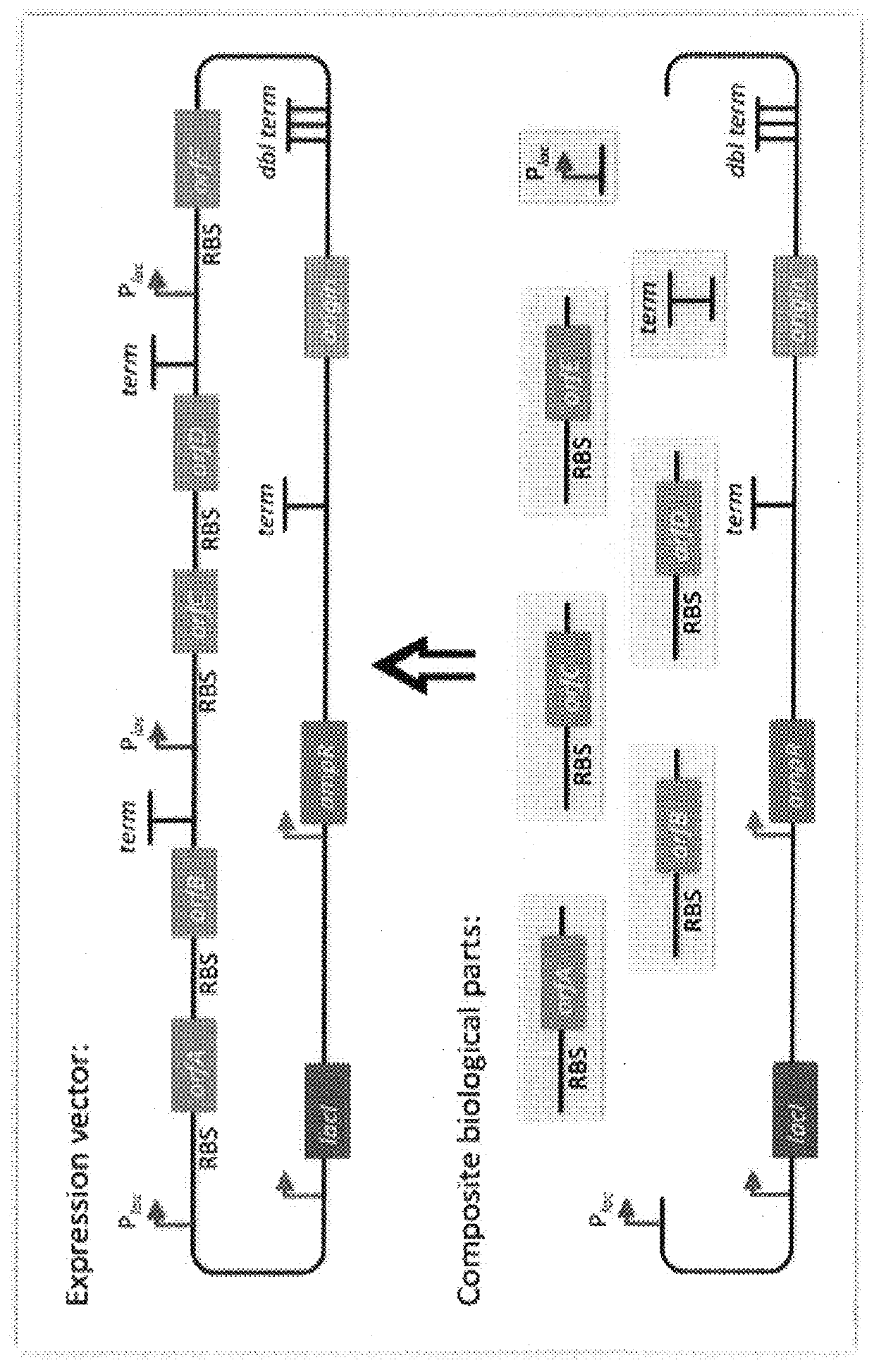
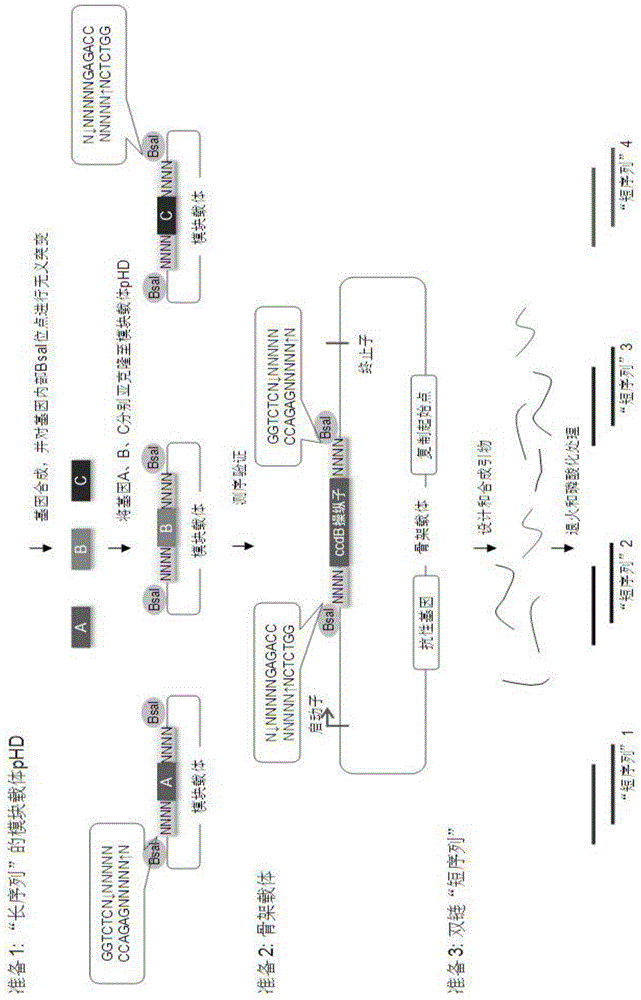
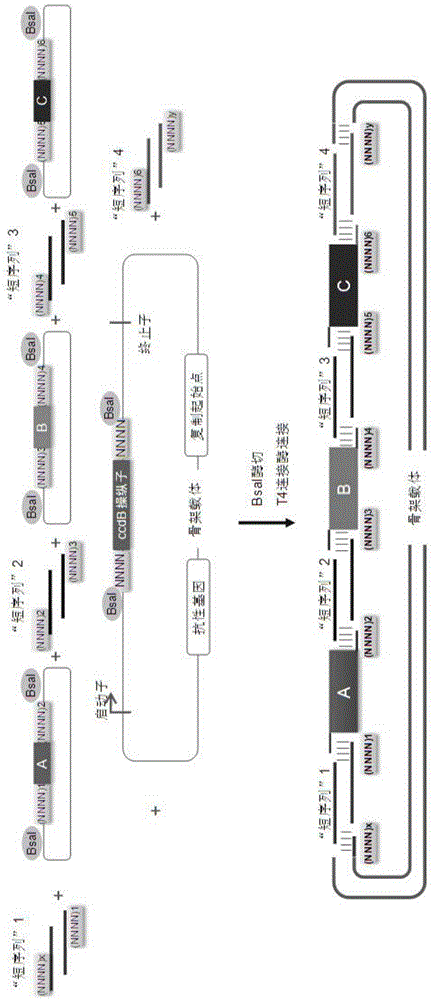
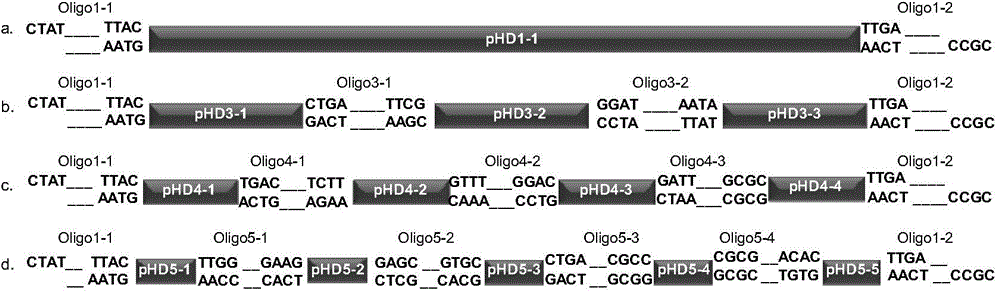
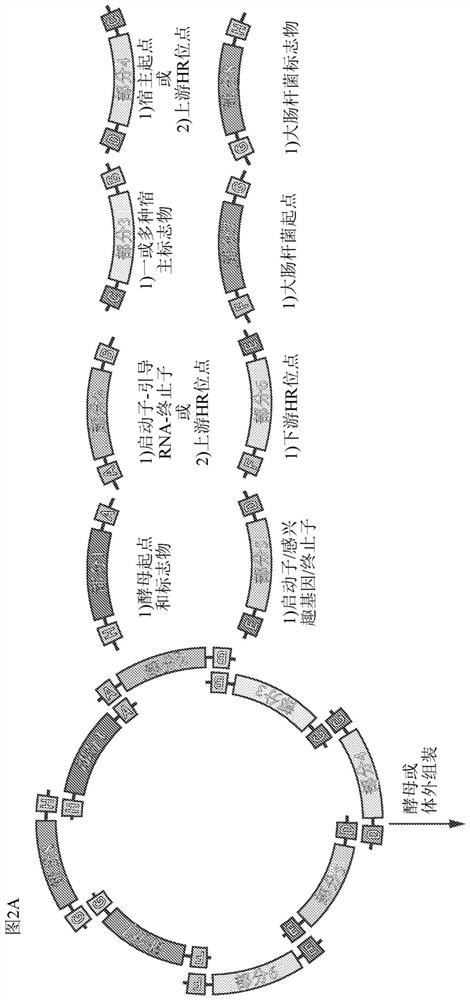
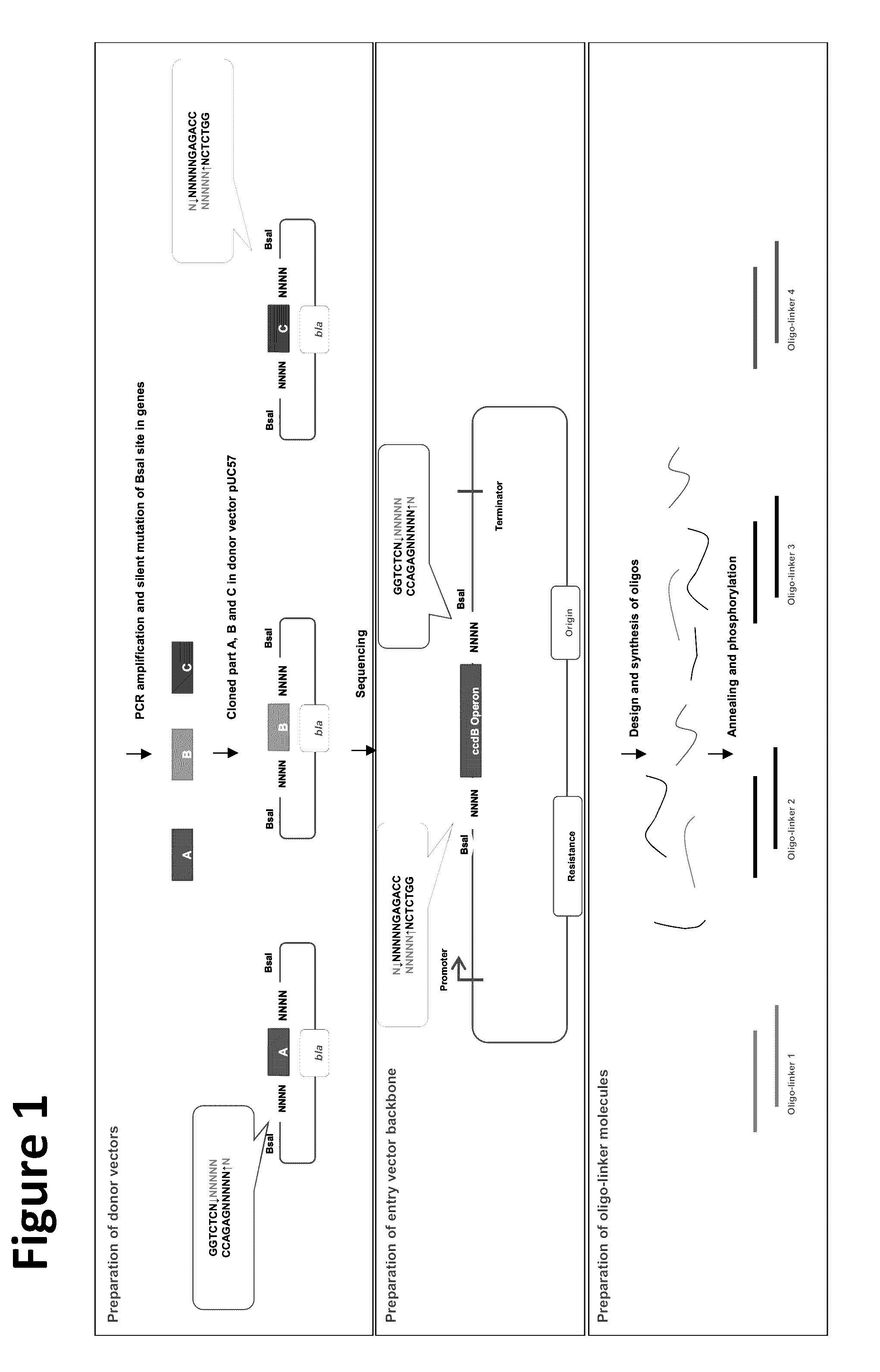
Scar-less multi-part DNA assembly design automation
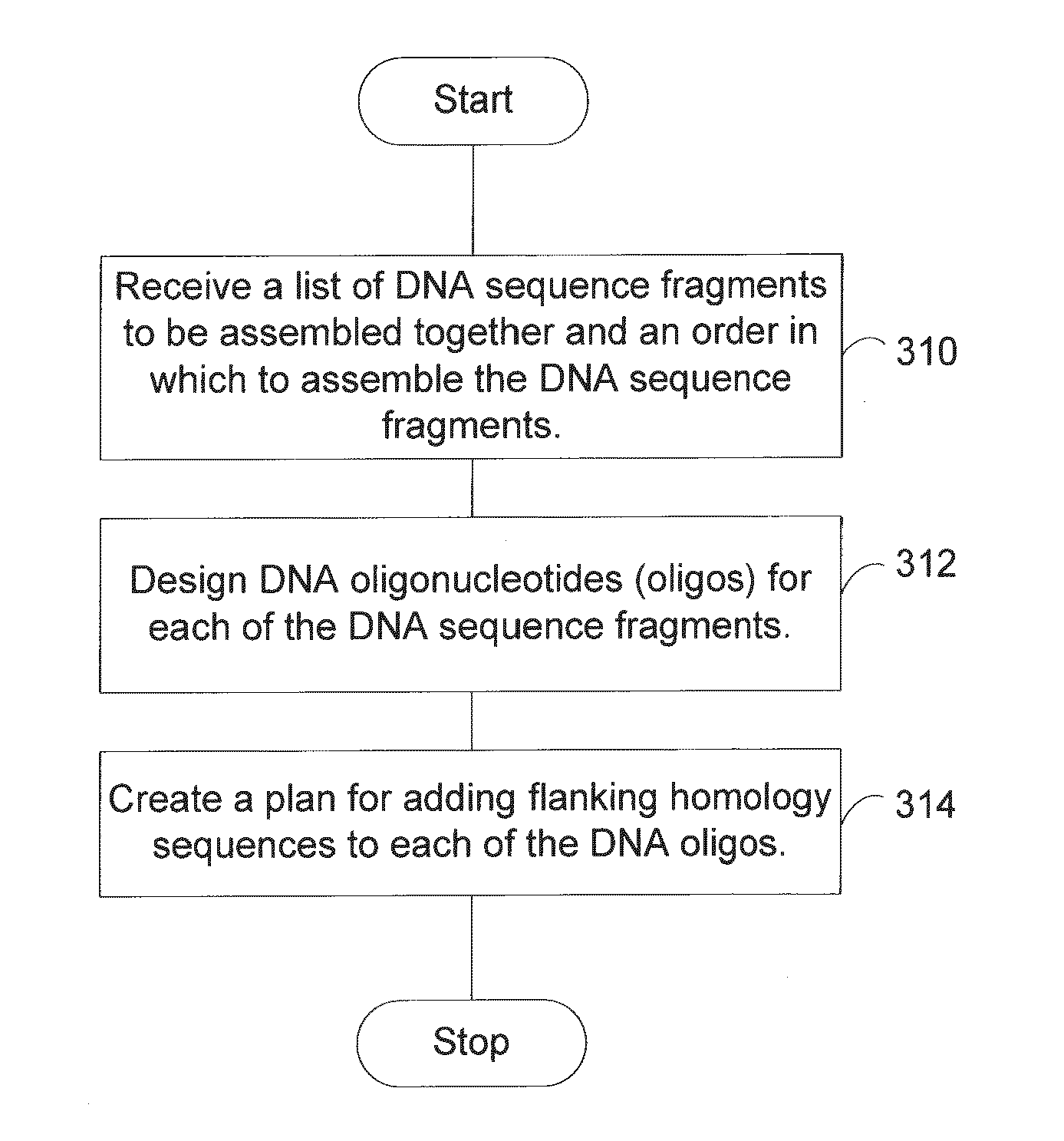
The present invention provides a method of a method of designing an implementation of a DNA assembly. In an exemplary embodiment, the method includes (1) receiving a list of DNA sequence fragments to be assembled together and an order in which to assemble the DNA sequence fragments, (2) designing DNA oligonucleotides (oligos) for each of the DNA sequence fragments, and (3) creating a plan for adding flanking homology sequences to each of the DNA oligos. In an exemplary embodiment, the method includes (1) receiving a list of DNA sequence fragments to be assembled together and an order in which to assemble the DNA sequence fragments, (2) designing DNA oligonucleotides (oligos) for each of the DNA sequence fragments, and (3) creating a plan for adding optimized overhang sequences to each of the DNA oligos.
Owner:RGT UNIV OF CALIFORNIA
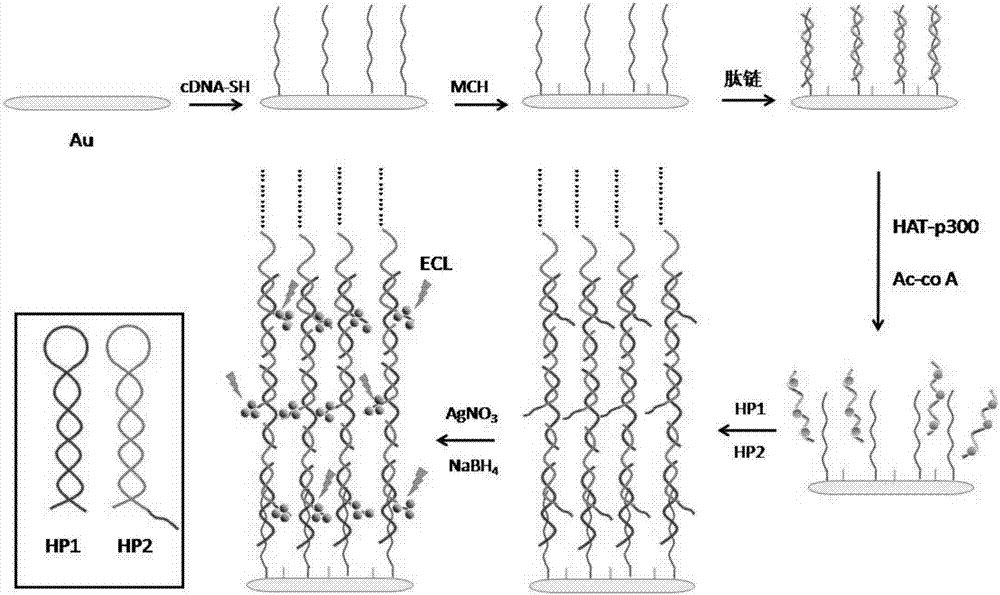
Construction of electrochemical luminescence sensor for detecting acetyltransferase activity
ActiveCN107101997AEasy to prepareEasy to operateChemiluminescene/bioluminescenceMaterial analysis by electric/magnetic meansPhysical chemistryAcetyltransferase activity
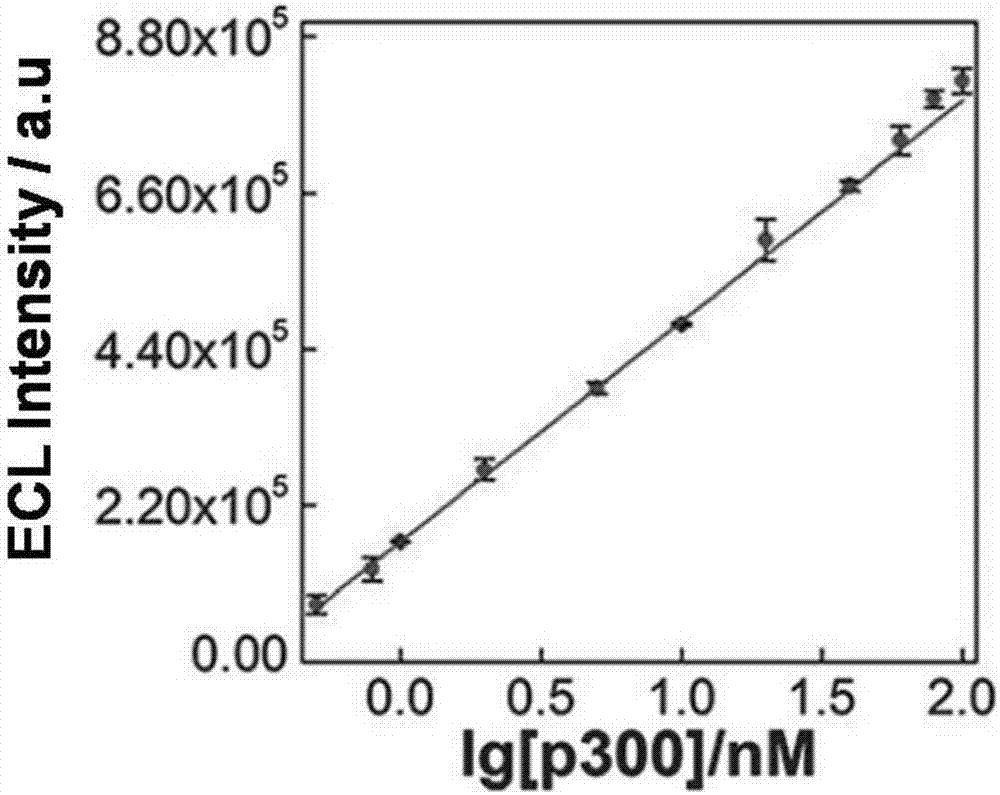
The invention relates to a construction of an electrochemical luminescence sensor for detecting acetyltransferase activity. The construction comprises the following steps: firstly, polishing, cleaning and activating a gold electrode, assembling captured DNA on the surface of the electrode through action of gold-sulfur bond, and enclosing non-specific binding sites on the surface of the electrode with MCH; then adsorbing a polypeptide chains through electrostatic action, performing acetylation treatment on the polypeptide chains under the action of HATp300, so as to separate from the surface of the electrode; secondly, performing hybrid chain reaction on the electrode in a hybrid chain solution containing two hairpin DNA, and reducing a silver cluster; and finally, detecting electrochemical luminescence signal of the assembled electrochemical luminescence sensor in the solution. The ECL biosensor has remarkable advantages on the quantitative analysis of HAT p300 and the activity analysis on HAT p300 in complicated cell lysis buffering solution.
Owner:青岛卓越海特信息技术有限公司
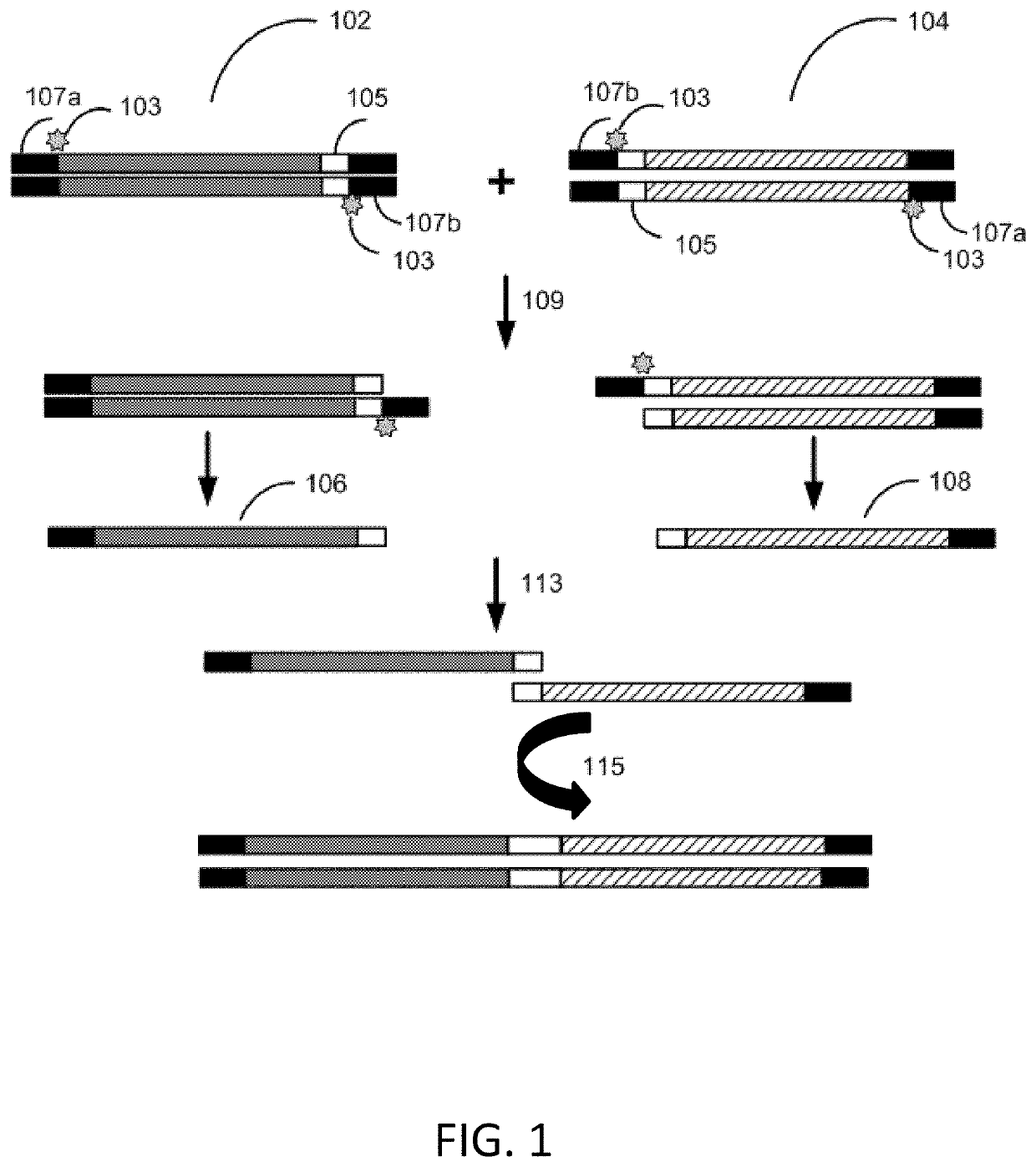
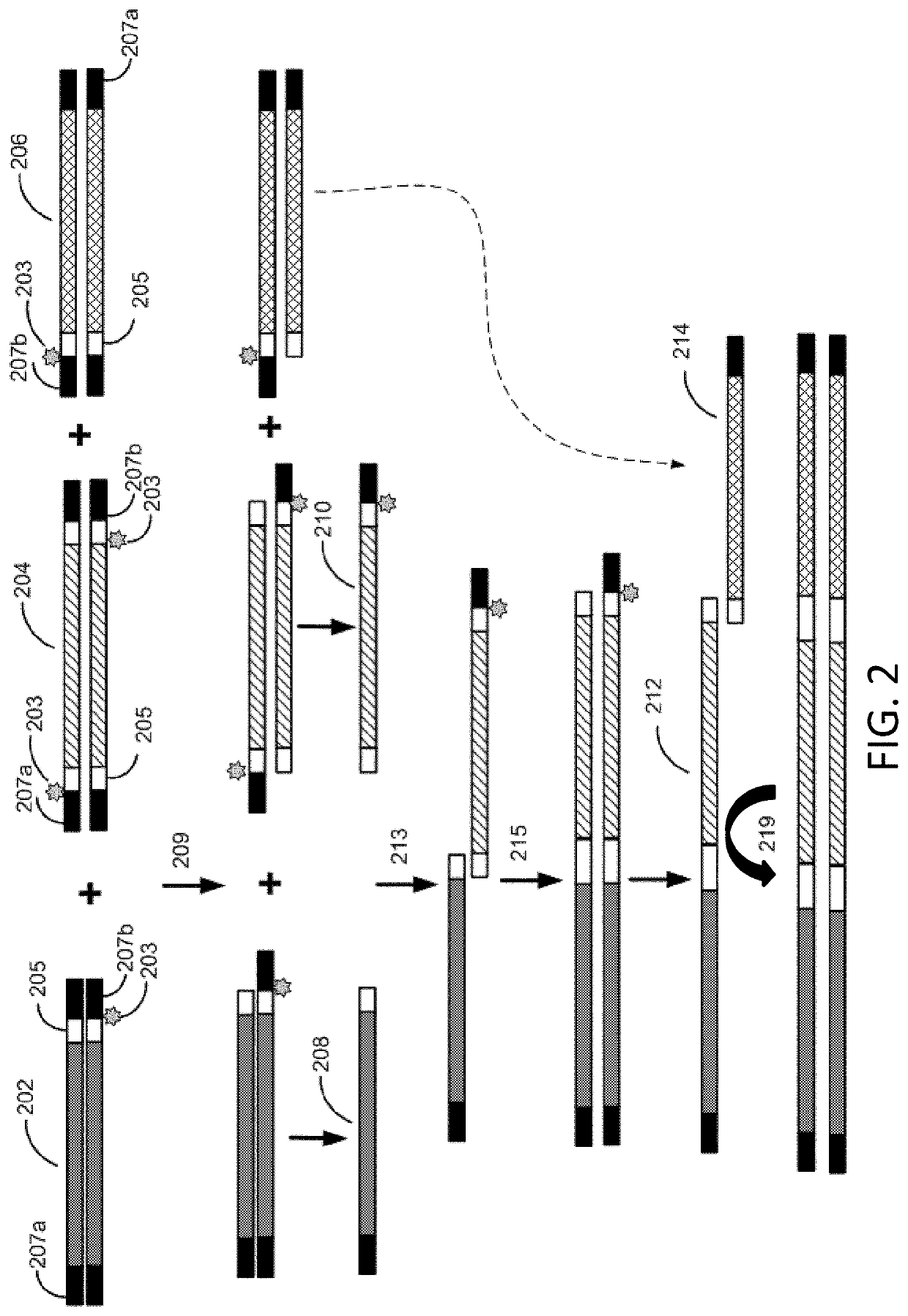
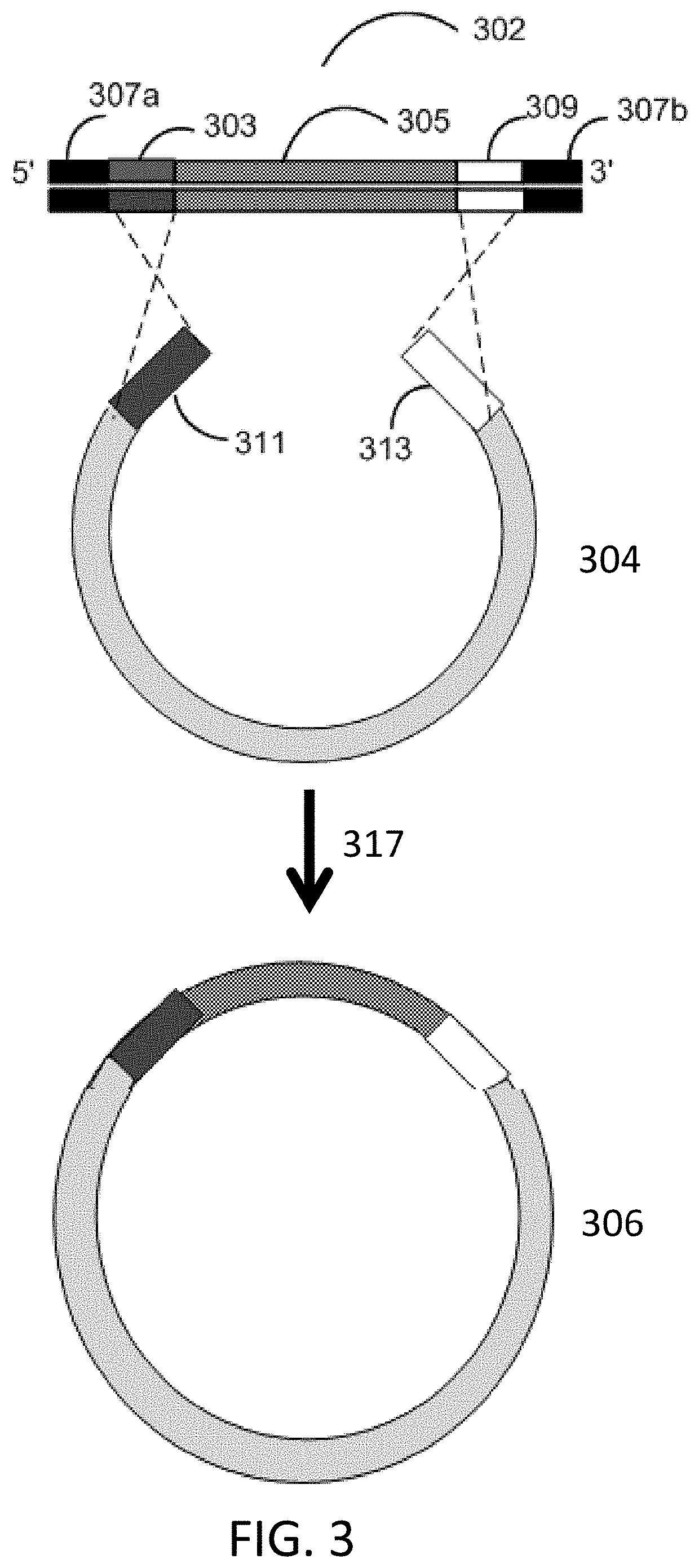
Methods for seamless nucleic acid assembly
Provided herein are methods, systems, and compositions for seamless nucleic acid assembly. Such methods, systems, and compositions for seamless nucleic acid assembly include those for in vitro recombination cloning, single-stranded hierarchal DNA assembly, or overlap extension PCR without primer removal.
Owner:TWIST BIOSCI
Adenoviral vector system and recombinant adenovirus construction method
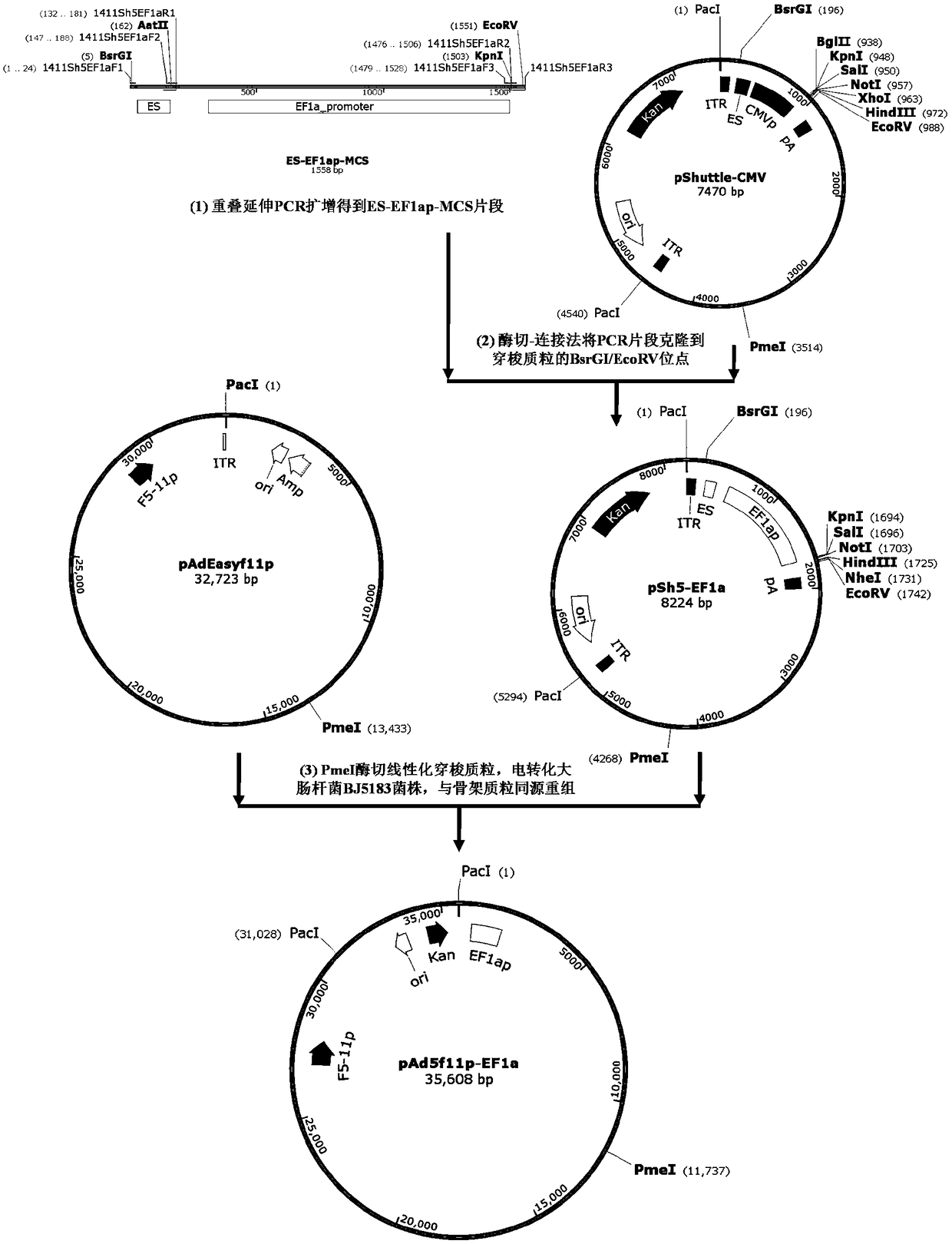
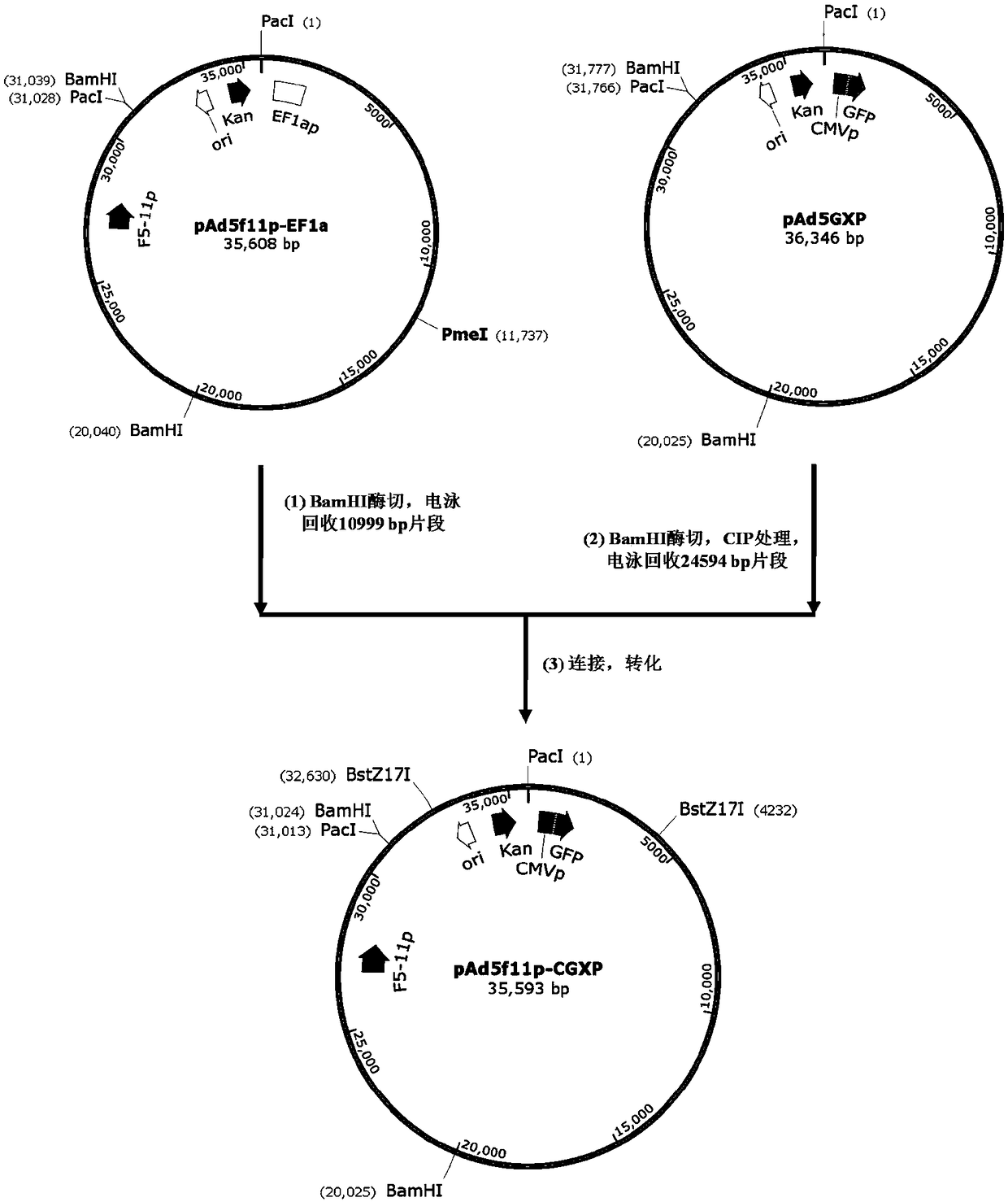
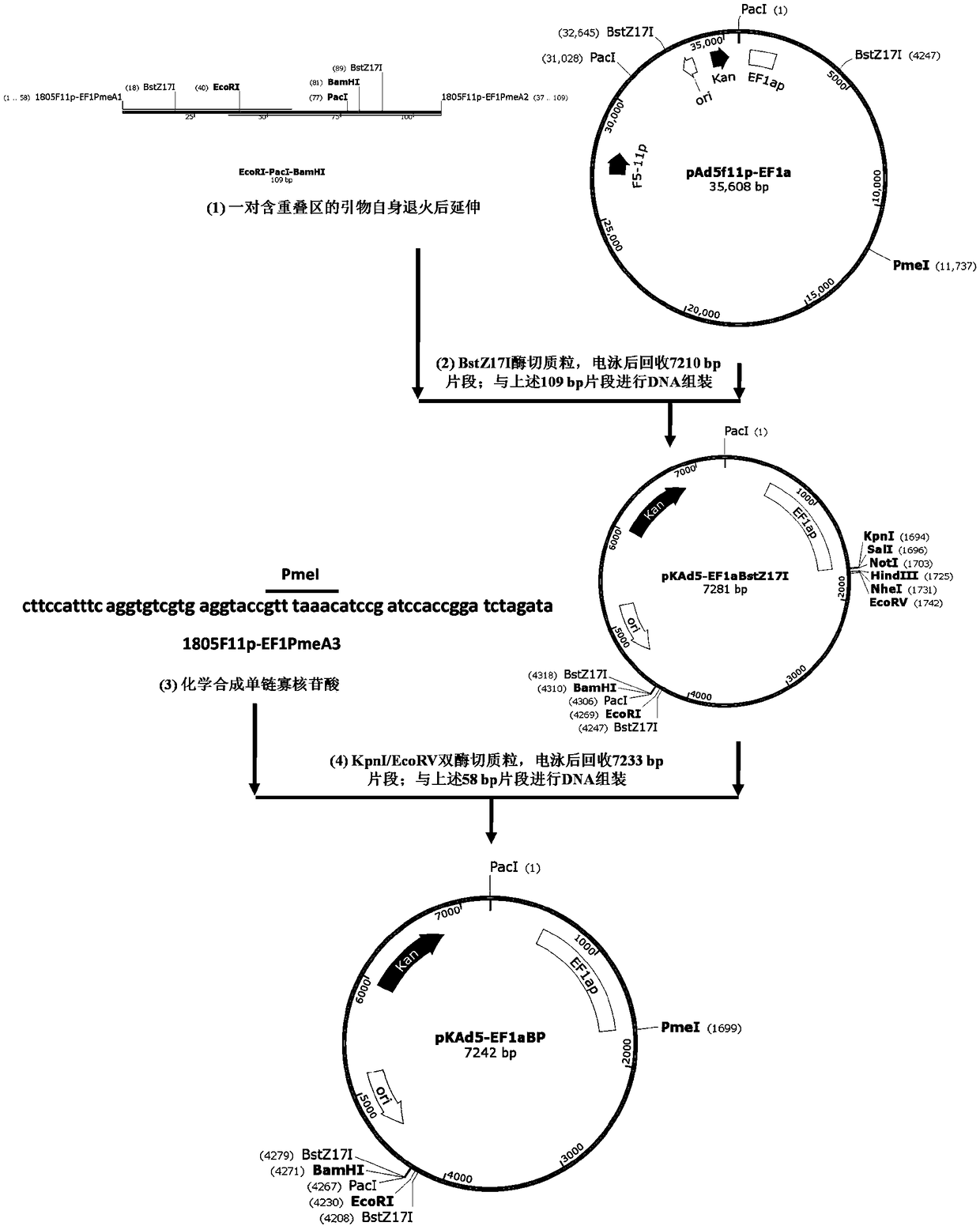
The invention provides an adenoviral vector system and a recombinant adenovirus construction method. The vector system comprises adenoviral plasmids pKAd5f11p-EF1aP and pKAd5f11pES-PmeI and shuttle plasmids pUC19-PM. The adenoviral plasmid contains an E1 / E3 deleted human adenovirus type 5 (HAdV-5) genome and the original HAdV-5 fiber gene is replaced by a fusion gene F5-11p of HAdV-5 and HAdV-11p.A PmeI site is an exogenous gene insertion site. Plasmids pKAd5f11p-EF1aP contain a human EF1a promoter in the original E1 region. The shuttle plasmids pUC19-PM are matched with the plasmids pKAd5f11pES-PmeI. The recombinant adenovirus construction method comprises: carrying out PCR amplification to obtain a desired gene fragment containing homologous overlapping regions on both sides and carrying out DNA assembling on the desired gene fragment and PmeI-linearized adenoviral plasmids to obtain adenovirus plasmids containing the desired exogenous gene, or cloning the multiple gene fragments tothe shuttle plasmids, shearing all the desired gene fragments through a restriction endonuclease and carrying out DNA assembling on the desired gene fragments and PmeI linearized pKAd5f11pES-PmeI toobtain the adenovirus plasmids containing the desired exogenous gene.
Owner:中国疾病预防控制中心病毒病预防控制所 +1
AC Impedance-Type DNA Electrochemical Sensor Based on Probe DNA Controlled Assembly Interface
InactiveCN102286371ALower impedanceSlow transfer rateBioreactor/fermenter combinationsBiological substance pretreatmentsBovine serum albuminDna assembly
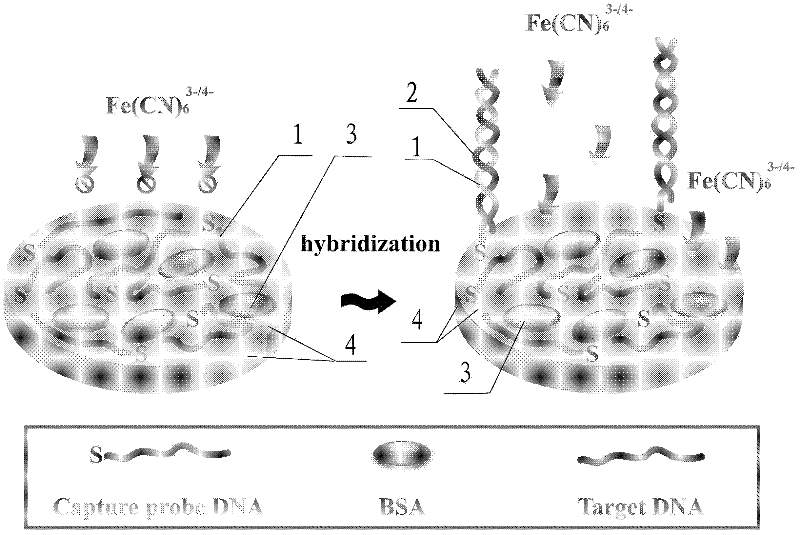
The invention discloses an alternating current impedance type deoxyribonucleic acid (DNA) electrochemical sensor based on a probe DNA control assembly interface, which comprises an electrode and capture probe DNA, wherein the electrode adopts a gold electrode, and the capture probe DNA adopts sulfydryl modified DNA. The alternating current impedance type DNA electrochemical sensor is characterized in that the sulfydryl modified DNA is modified onto the surface of the gold electrode through the chemical bonding effect of Au-S bonds with the gold electrode at the room temperature and the absorption effect of the probe basic group parts and the gold so that the capture probe DNA flatly lies on the surface of the gold electrode for forming a capture probe DNA assembly layer, and bovine serum albumin exists on the surface of the capture probe DNA assembly layer and is used as sealing agents and protection agents. The impedance change before and after the hybridization is used as indicationsignals, the method adopts the surface assembly chemical technology for building the flat lying type DNA probe identification interface, simultaneously, the form of the probe maintains the flat lyingstate on the surface of the electrode without being influenced by the blank hybrid condition, and the method has the characteristics of high sensitivity, good specificity and the like.
Owner:FUJIAN MEDICAL UNIV
Scar-Less Multi-Part DNA Assembly Design Automation
ActiveUS20120259607A1Analogue computers for chemical processesProteomicsDna assemblyComputational biology
The present invention provides a method of a method of designing an implementation of a DNA assembly. In an exemplary embodiment, the method includes (1) receiving a list of DNA sequence fragments to be assembled together and an order in which to assemble the DNA sequence fragments. (2) designing DNA oligonucleotides (oligos) for each of the DNA sequence fragments, and (3) creating a plan for adding flanking homology sequences to each of the DNA oligos. In an exemplary embodiment, the method includes (1) receiving a list of DNA sequence fragments to be assembled together and an order in which to assemble the DNA sequence fragments, (2) designing DNA oligonucleotides (oligos) for each of the DNA sequence fragments, and (3) creating a plan for adding optimized overhang sequences to each of the DNA oligos.
Owner:RGT UNIV OF CALIFORNIA
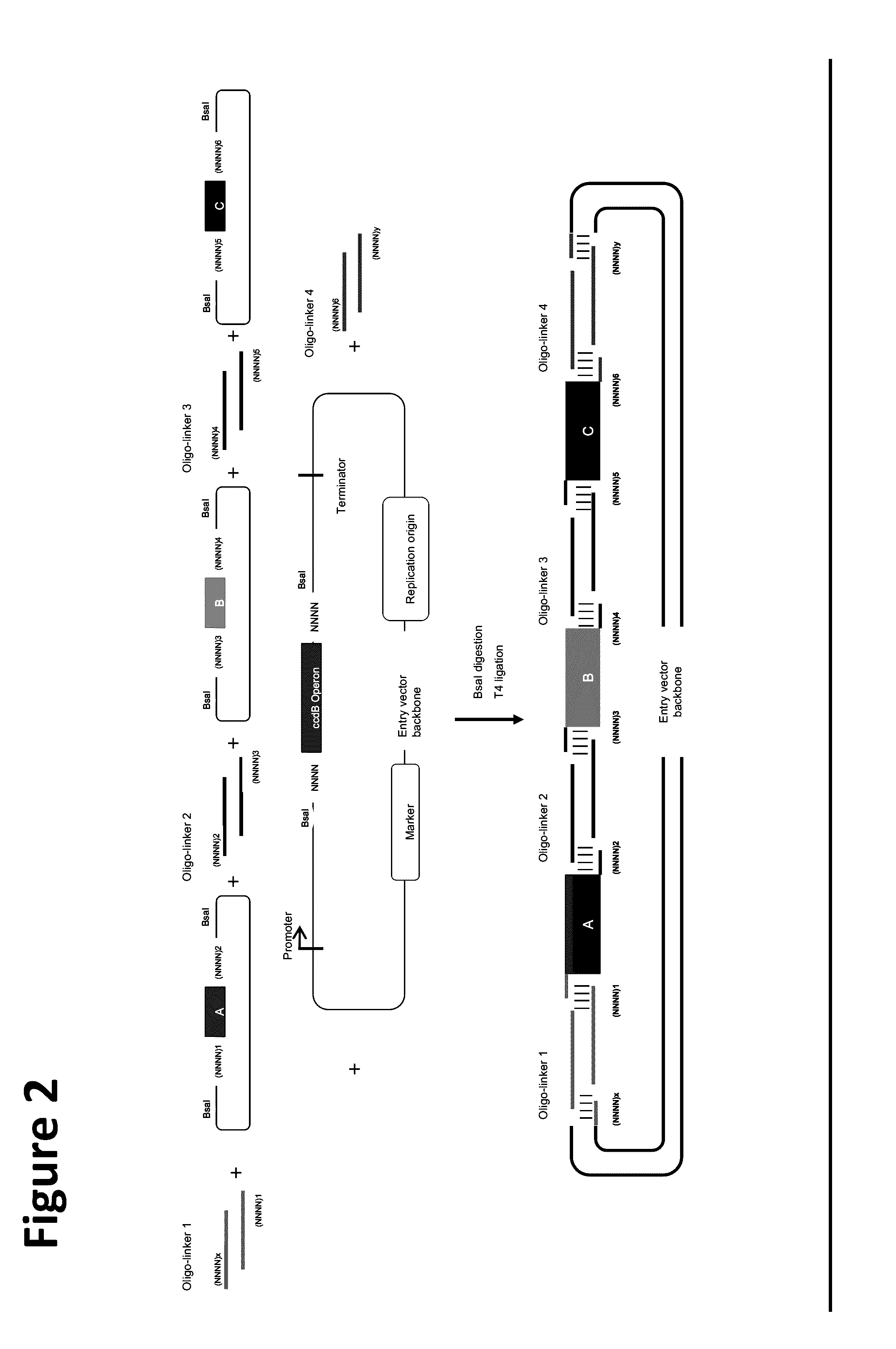
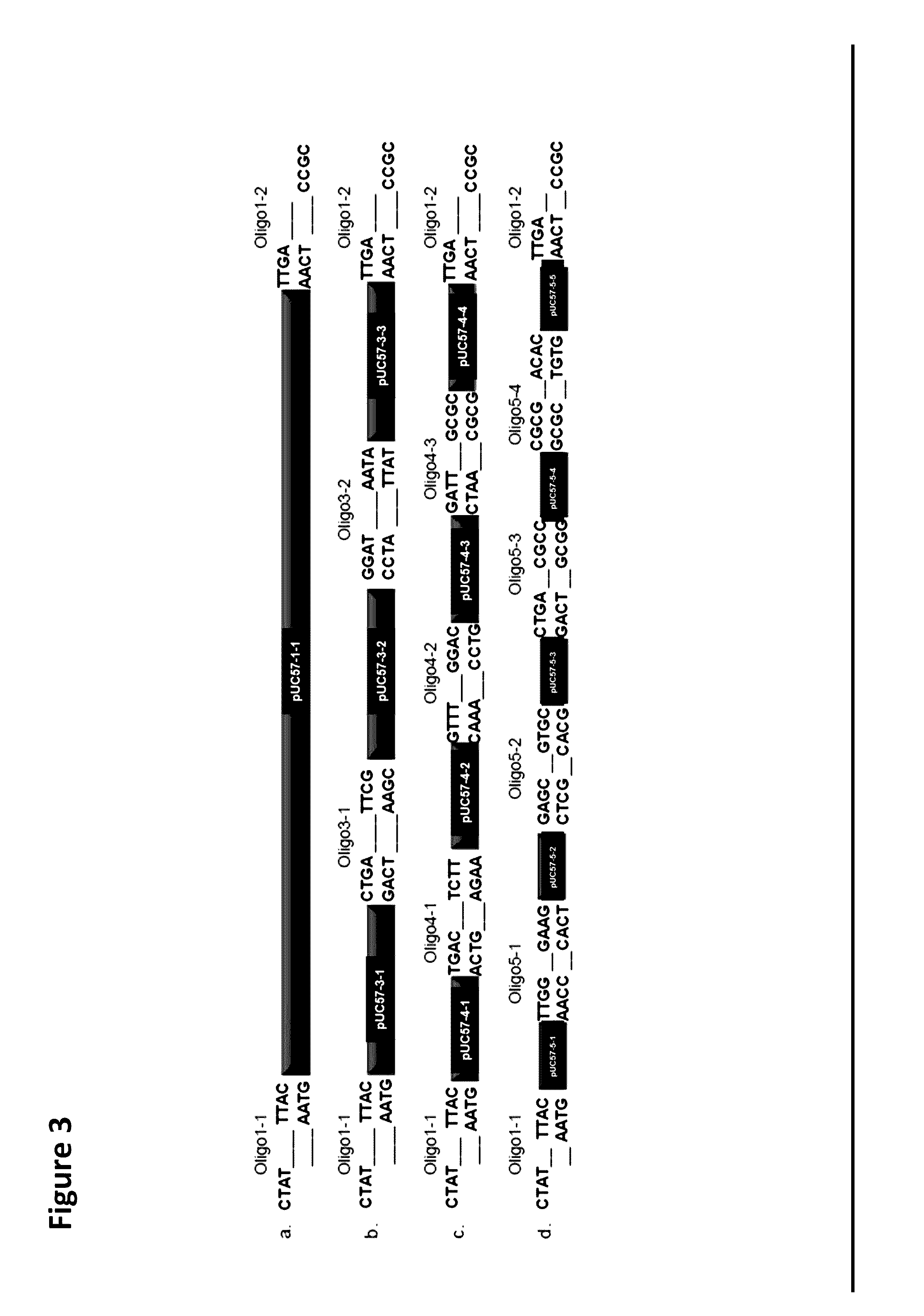
Oligonucleotide-linker-mediated DNA assembly method and application thereof
ActiveCN106282157ATo achieve the purpose of flux optimization pathwayAvoid residueVector-based foreign material introductionDNA preparationOperonBarcode
The invention discloses an oligonucleotide-linker-mediated DNA assembly method and an application thereof. The assembly method comprises steps of synthesis of a long sequence, preparation and assembly of a module carrier, remolding and assembly of a skeleton carrier, preparation of a short sequence, and enzyme-cut and link-up integration of the module carrier, the remolded skeleton carrier and the short sequence. According to the method, the short sequence is a regulating element such as a promoter and an RBS, and the long sequence is a replication initiation point of a gene or a carrier. The short sequence is designed to synthesize a double-stranded DNA chain, and can connect the long sequence and be used as a barcode sequence to determine the assembly sequence in an assembly process. The gene sequence in an operon is achieved by synthesizing short sequence with different cohesive ends. According to the method, multiple factors are allowed to participate in gene regulation at the same time. The method can be used to construct a combinatorial library of a biosynthesis channel, and a high-flux optimized channel is achieved.
Owner:NANJING GENSCRIPT BIOTECH CO LTD
A Method for Parallel Assembly of Multiple Segments of DNA
InactiveCN102296059AImprove efficiencyAvoid pollutionMicroorganism based processesVector-based foreign material introductionBiological bodyExtracellular
The invention relates to a method for parallel assembly of multi-segment DNA. Choose to insert the circular or linear unit vector into the circular host or linear host to form a gene carrier, and then recover the circular or linear vector to obtain the recombined DNA genetic material. Its characteristic is that it can realize parallel and simultaneous operation of multiple pieces of DNA, thereby improving the efficiency of DNA assembly to form a new DNA structure, and preventing DNA pollution and disproportionation, ensuring the quality of assembled DNA, so that it can achieve the design purpose. The time required to assemble multiple pieces of DNA is approximately proportional to log(N). The assembly operation is carried out outside or inside the cell, so that multiple DNA molecules can be spliced in any order to form a new DNA structure. It is suitable for application in the field of genetic engineering to obtain new organisms through genetic recombination.
Owner:石振宇
Method for rna-guided endonuclease-based DNA assembly
InactiveUS20180105806A1Low efficiencyLow dissociation rateHydrolasesTransferasesSingle strandLigation
The invention provides a novel approach to facilitate assembly of DNA molecules. This approach utilizes RNA-guided endonucleases, capable of targeting any DNA sequence, which cleave DNA and generate DNA fragments characterized by single stranded overhangs. After annealing of complementary overhangs, DNA fragments are covalently connected, generating a single DNA molecule. In this way, the present invention combines the reliability of classic restriction-ligation techniques, removes all sequence constraints from the desired final DNA molecule, and expands the number of DNA pieces that can be assembled at once.
Owner:MASSACHUSETTS INST OF TECH
Nuclease-mediated DNA assembly
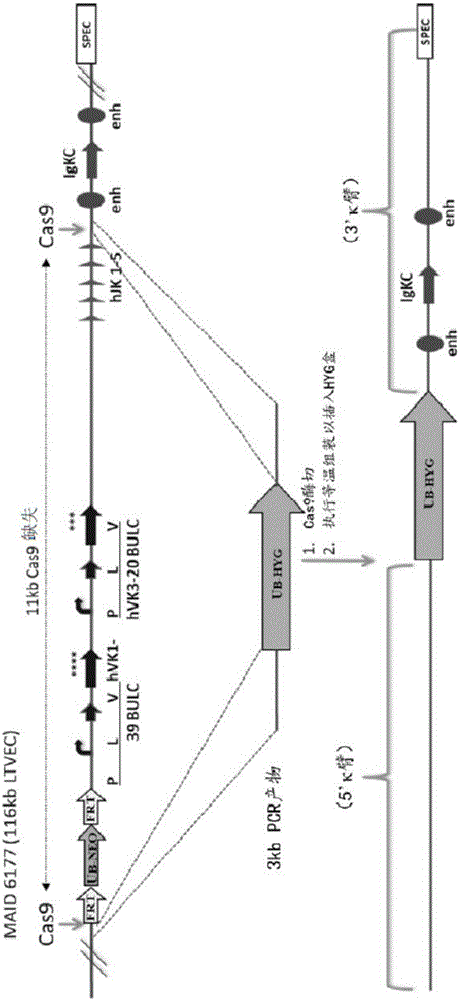
Methods are provided herein for assembling at least two nucleic acids using a sequence specific nuclease agent (e.g., a gRNA-Cas complex) to create end sequences having complementarity and subsequently assembling the overlapping complementary sequences. The nuclease agent (e.g., a gRNA-Cas complex) can create double strand breaks in dsDNA in order to create overlapping end sequences or can create nicks on each strand to produce complementary overhanging end sequences. Assembly using the method described herein can assemble any nucleic acids having overlapping sequences or can use a joiner oligo to assemble sequences without complementary ends.
Owner:REGENERON PHARM INC
Multi-adenine based DNA capture probe, biosensor and detection method thereof
ActiveCN106338539AAssembly density can be adjustedRegulating assembly densityMaterial analysis by electric/magnetic meansModified dnaElectrochemical biosensor
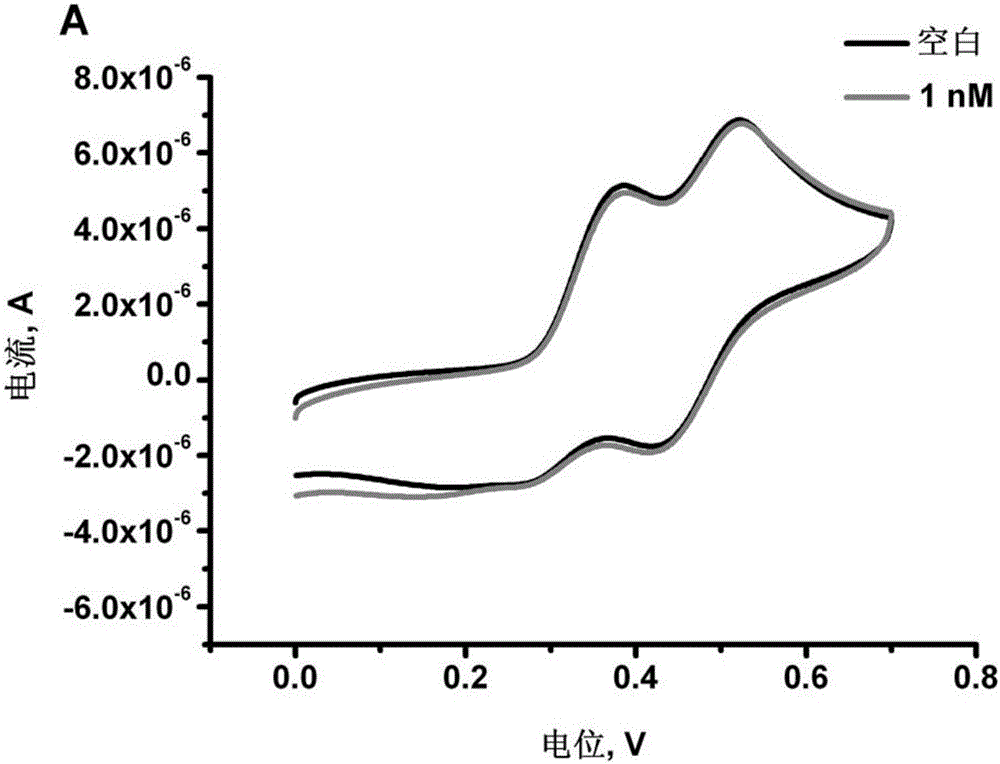
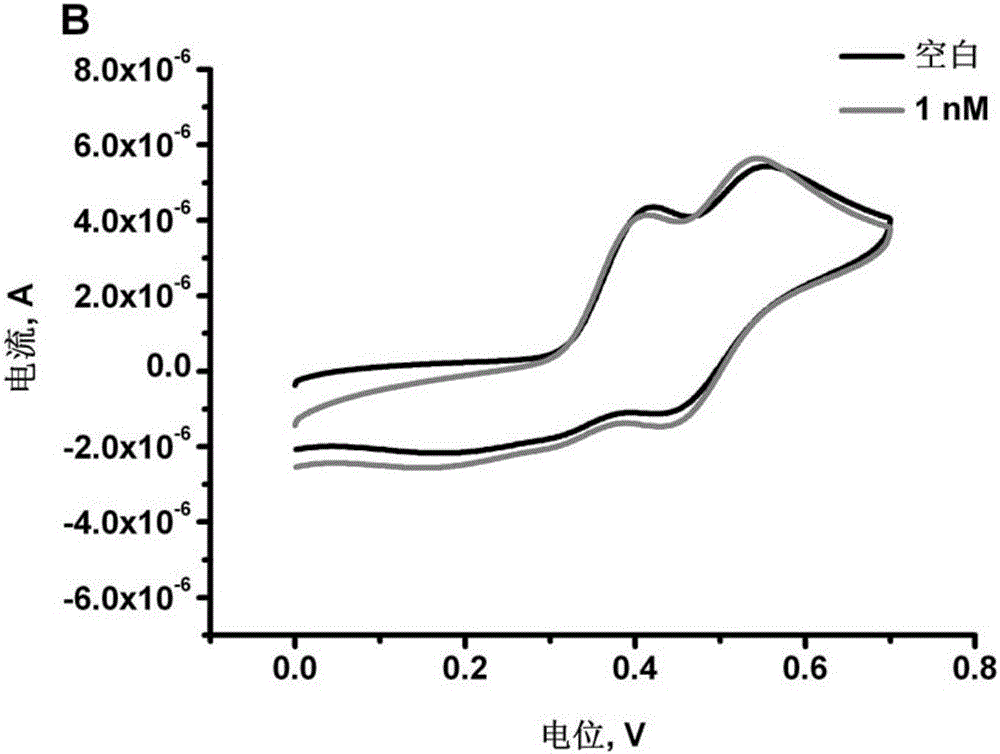
The invention discloses amulti-adenine based DNA capture probe, a biosensor and a detection method thereof. The nucleotide sequence of the DNA capture probe is shown as a sequence table SEQID No.1, SEQID No.2, SEQID No.3, SEQID No.4 or SEQID No.5. The mutual effect of adenines and a gold electrode to fix the multiple-consecutive-adenine DNA probe to the surface of the gold electrode, the electrochemical biosensor based on multi-adenine DNA is established for the first time, the effect same as a sulfydryl modified DNA based assembled gold electrode can be achieved, and the new era of novel electrochemical biosensors is opened. The multi-adenine based DNA capture probe can detect DNA in a high sensitivity and high specificity mode and is easy and convenient to operate and wide in application prospect, and the sensitivity can be up to 1 pM.
Owner:SHANGHAI INST OF MEASUREMENT & TESTING TECH
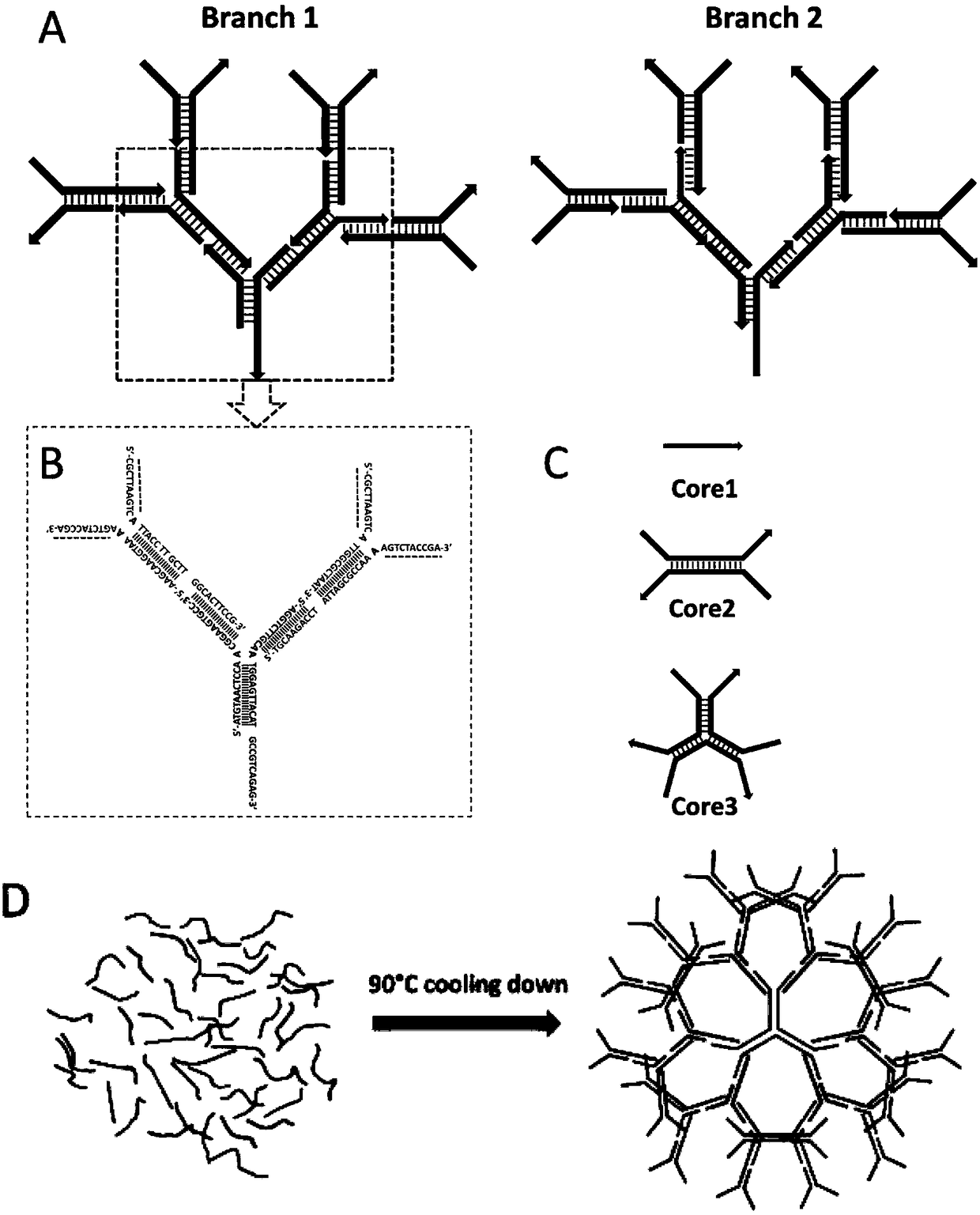
Preparation method and application of dendritic DNA assembly
ActiveCN108611348APrecise and controllable structureImprove flexibilityPharmaceutical non-active ingredientsDNA/RNA fragmentationMedicineSingle strand
The invention provides a preparation method and application of a dendritic DNA assembly. The preparation method of the dendritic DNA assembly includes the steps of: mixing several single stranded DNAin a buffer solution, performing heating to 90DEG C, and then conducting cooling to 4DEG C at a rate not exceeding 3.6DEG C / min, thus obtaining the dendritic DNA assembly. The invention also providesapplication of the dendritic DNA assembly as a molecular carrier in imaging, diagnosis and treatment. The invention has the advantages that: through one-step quenching treatment of a single stranded DNA mixture, the dendritic DNA assembly with precisely controllable structure can be obtained.
Owner:SHANGHAI NINTH PEOPLES HOSPITAL SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
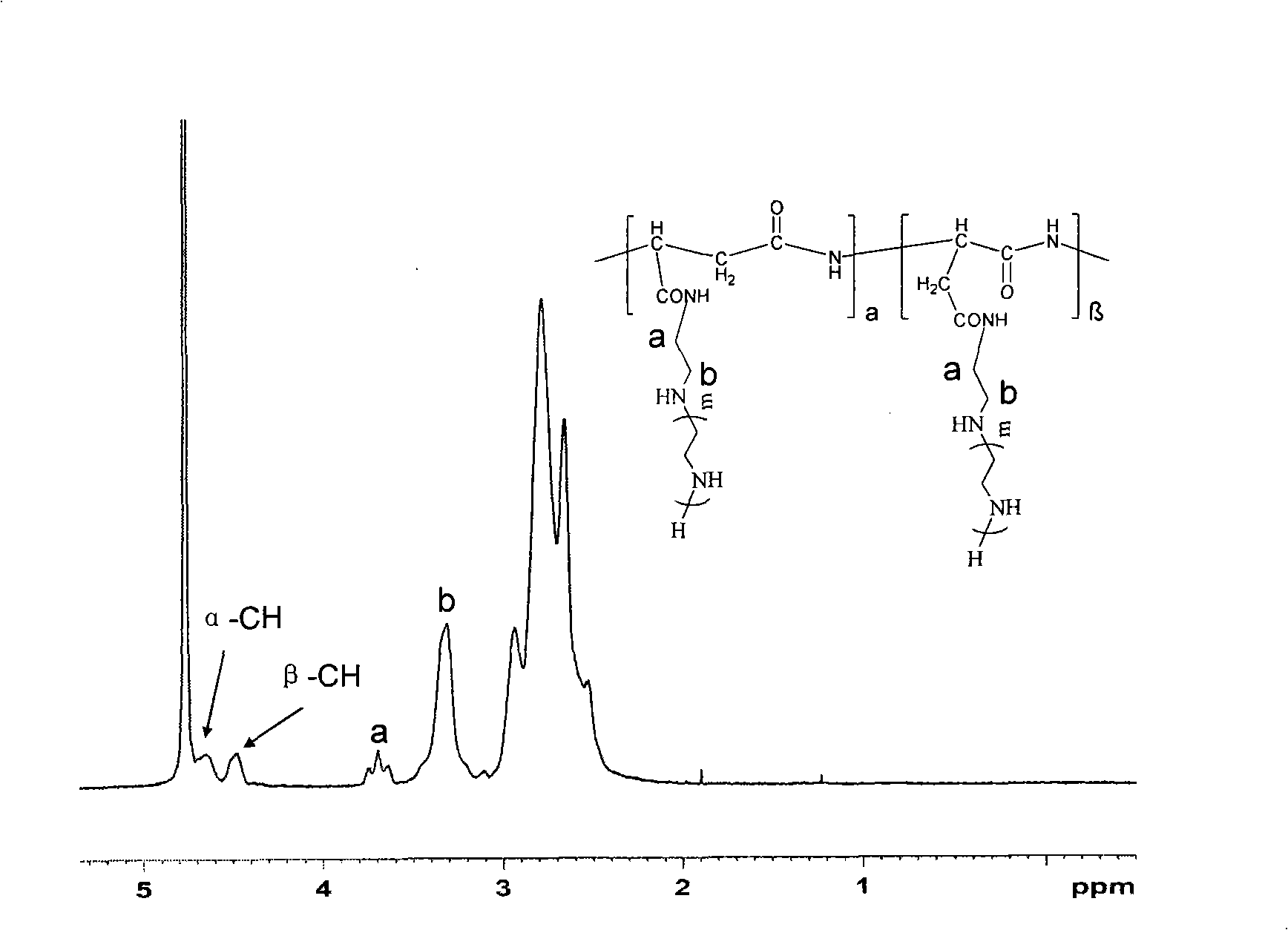
Preparation and application of amphiphilic polypeptide genetic vector
ActiveCN108623804ALow toxicityEasy to controlOther foreign material introduction processesSynthesis methodsPlasmid dna
The invention discloses preparation and application of an amphiphilic polypeptide genetic vector, and belongs to the technical field of nanometer medical material preparation. The method comprises thefollowing steps of using hexamethyldisilazane as an initiator to initiate two kinds of polypeptide monomers of CBZ-L-lysine-NCA and L-leucine-NCA to perform NCA ring opening polymerization; finally removing CBZ protection groups to obtain an amphiphilic embedded polypeptide genetic vector of poly(L-lysine)50-b-poly(L-leucine)15. The polypeptide polymer provided by the invention has low toxicity andcan be used for compressing plasmid DNA to assemble regular nanometer particles; the transfection efficiency is high; the vector belongs to an effective genetic vector; in addition, the controllablepolymerization is used by the synthesis method; natural polypeptide is used as raw materials; the obtained polymers are safe and efficient.
Owner:JIANGSU UNIV
Method for rapidly assembling non-phosphorylated DNA (deoxyribonucleic acid) fragments in vitro
ActiveCN105950613AIncrease costHigh throughput operationMicrobiological testing/measurementDNA preparationEnzyme GeneDNA fragmentation
The invention discloses a method for rapidly assembling non-phosphorylated DNA (deoxyribonucleic acid) fragments in vitro, and belongs to the technical field of genetic engineering. Homologous arm sequences are introduced to the two terminals of a target fragment to be assembled and constructed, and the DNA fragment is rapidly assembled and constructed in vitro in a manner of thermal cycling for modification, annealing, cutting and connection. 2 to 4 non-phosphorylated DNA fragments can be efficiently, rapidly and seamlessly assembled at one time within 1.5h. The method can be used for conventional DNA recombination and construction and implementing high-throughput operation, and is particularly suitable for in-vitro efficient assembling of enzyme genes and synthetic pathways, and rich and diversified combinations and mutants can be introduced for rapid directional evolution and biological synthesis operation.
Owner:JIANGNAN UNIV
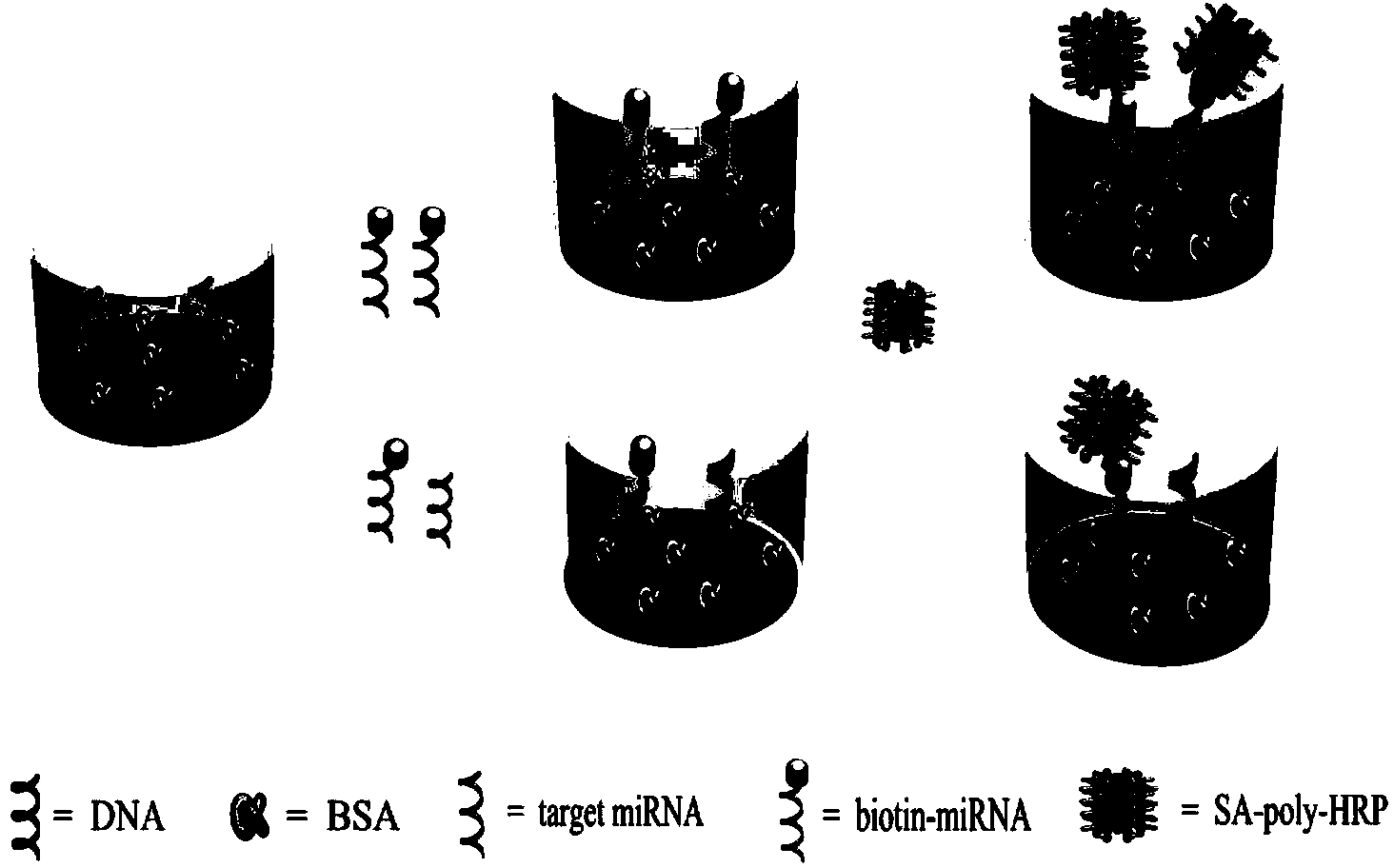
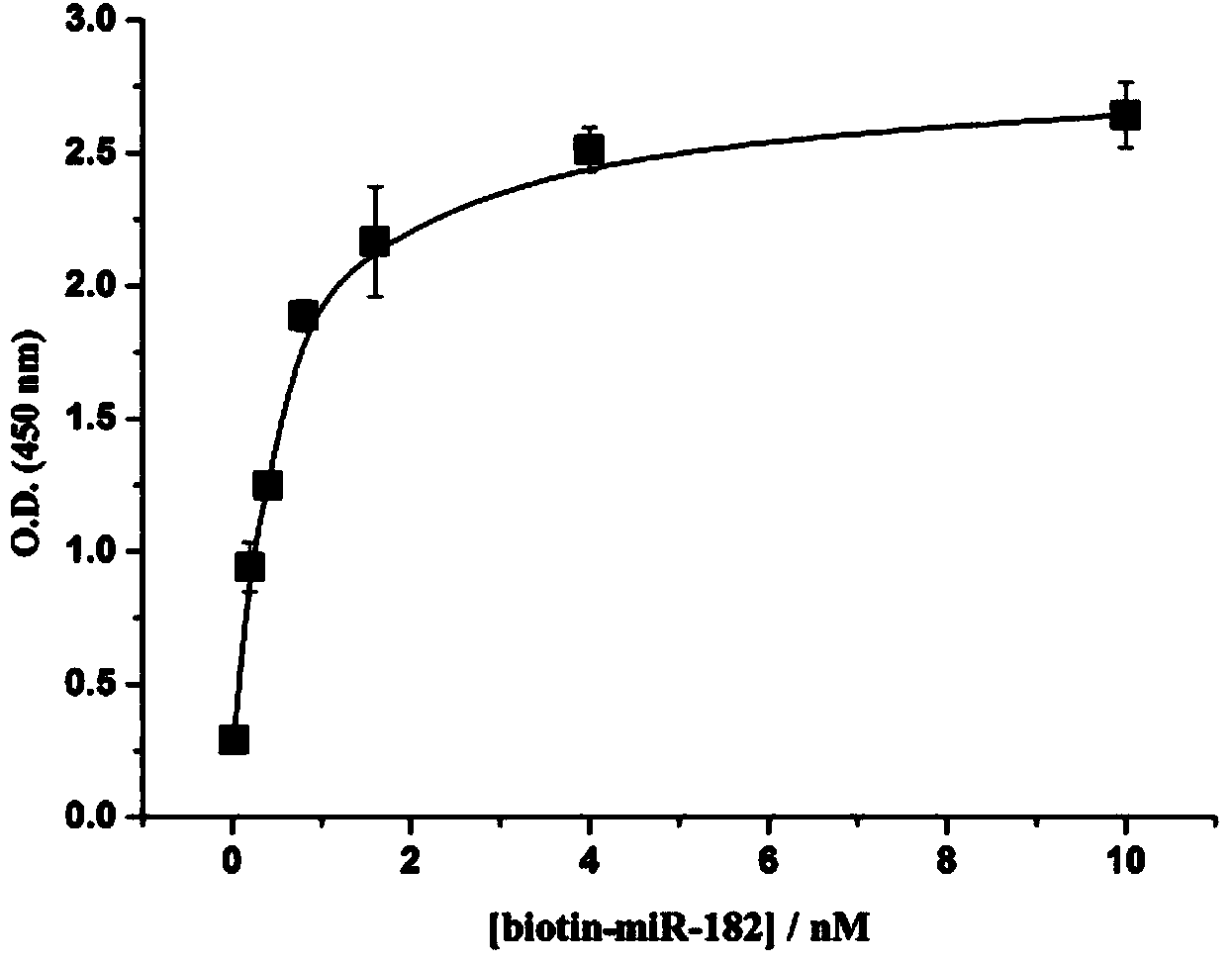
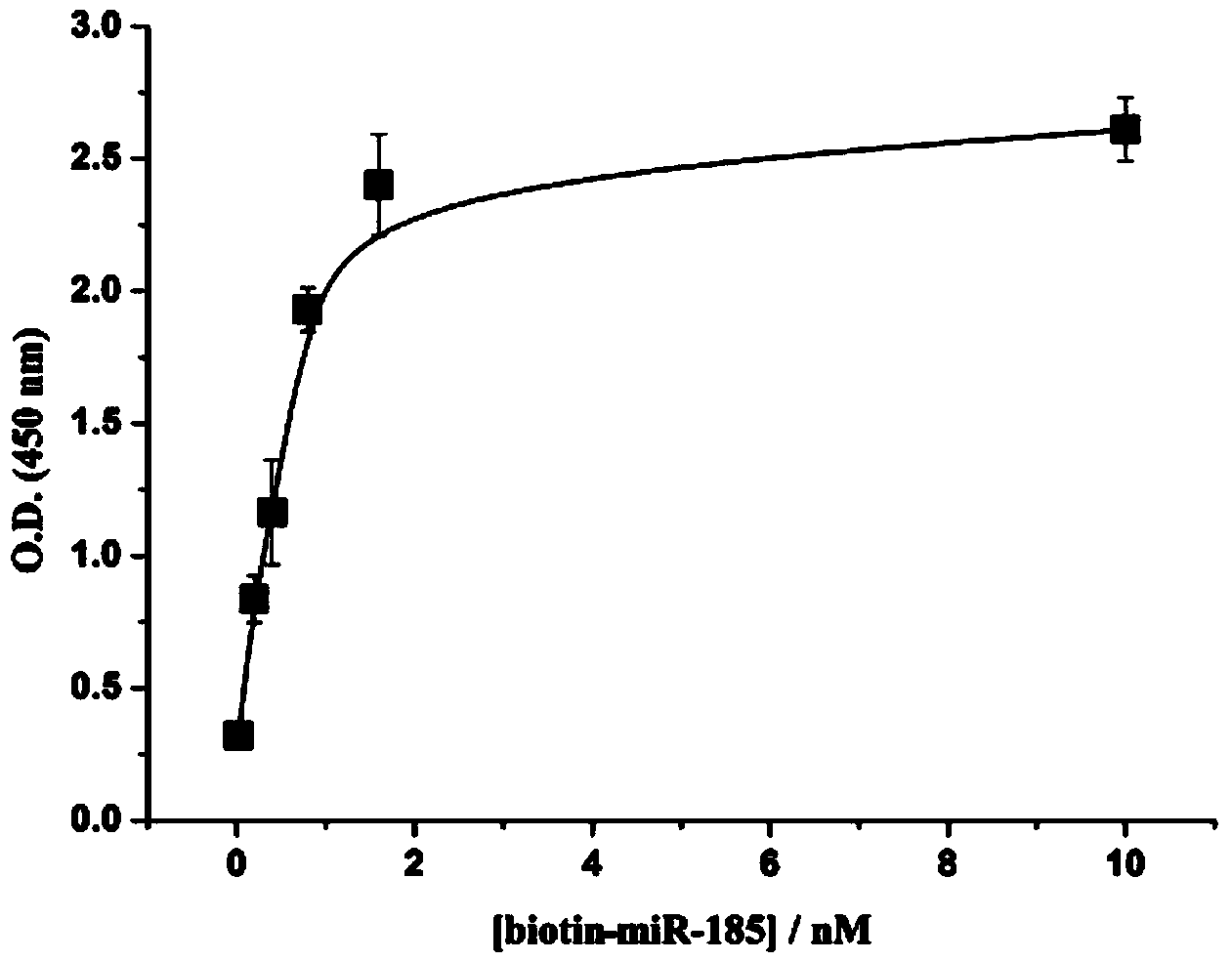
Colorimetric method for simultaneously detecting multiple miRNA (micro-ribonucleic acid) sequences based on competitive hybridization reaction
InactiveCN103529200AHigh sensitivitySimple methodMicrobiological testing/measurementColor/spectral properties measurementsModified dnaHybridization reaction
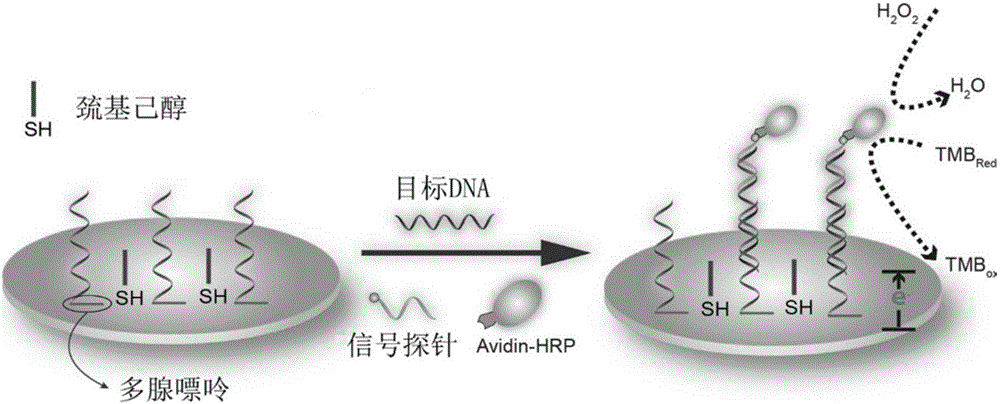
The invention discloses a colorimetric method for simultaneously detecting multiple miRNA (micro-ribonucleic acid) sequences based on a competitive hybridization reaction. The method comprises the following steps: assembling amino modified DNA (deoxyribonucleic acid) on the surface of a micropore plate, and closing micropores; performing a competitive hybridization reaction on biotin modified miRNA and a target miRNA sequence with a fixed DNA probe on the surface of the micropores; deriving avidin marked poly-horseradish peroxidase to the surface of the micropores, and catalyzing a chromogenic reaction between TMB (tetramethylbenzidine) and hydrogen peroxide by the poly-horseradish peroxidase. The target miRNA sequence is detected by determining the absorbance of a colored solution. The method is simple, quick, high in sensitivity and good in selectivity, does not need to mark an actual sample, and can be directly used for detecting multiple miRNA sequences.
Owner:CENT SOUTH UNIV
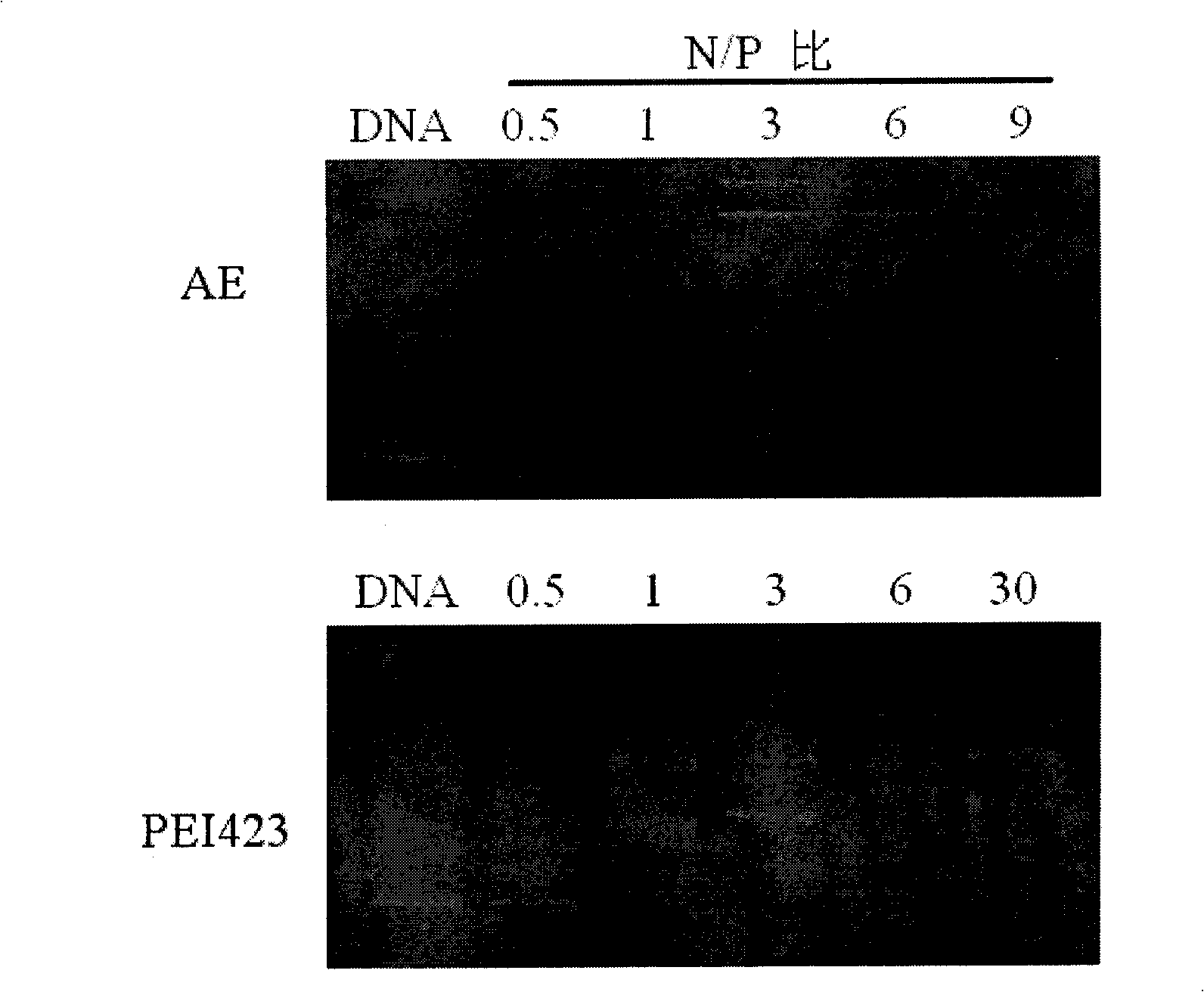
Assembly method for core-shell structural gene support system and uses thereof
InactiveCN101492691AThe synthesis method is simpleLow costVector-based foreign material introductionBlood plasmaDna assembly
The invention discloses a method for assembling a gene vector system with a nuclear shell structure. The method has the following steps: complete ring-opening is carried out on small molecular weight polymine (PEI) to gather L-aspartame (PSI), alpha, beta-polyL-aspartame-g-polymine (AE) are obtained after purification, the AE is taken as a carrier to be assembled into a nano gene vector system with the nuclear shell structure with DNA; the invention is characterized in that the synthetic method of brush-shaped polycation is simple and the cost is low; as the used raw material is free from toxicum, the gene vector of the obtained brush-shaped polycation has low toxicity; the brush-shaped polycation and DNA can form nano-particle with the nuclear shell structure by charge interaction; the nano-particle has high gene transfection efficiency; base on a hydrophilic shell layer, the nano-particle can reduce absorption on protein, so that the nano-gene vector system with the nuclear shell structure has fine gene transfection efficiency with the existence of blood plasma and is expected to be used in treatment of in vivo genes.
Owner:EAST CHINA NORMAL UNIV
APPLICATIONS OF CRISPRi IN HIGH THROUGHPUT METABOLIC ENGINEERING
The disclosure describes methods, compositions, and kits for high throughput DNA assembly reactions in vitro. The disclosure further describes modular CRISPR DNA constructs comprising modular insert DNA parts flanked by cloning tag segments comprising pre-validated CRISPR protospacer / protospacer adjacent motif sequence combinations. High throughput methods of CRISPRi and CRISPRa are also disclosed.
Owner:ZYMERGEN INC
Methods for seamless nucleic acid assembly
Provided herein are methods, systems, and compositions for seamless nucleic acid assembly. Such methods, systems, and compositions for seamless nucleic acid assembly include those for in vitro recombination cloning, single-stranded hierarchal DNA assembly, or overlap extension PCR without primer removal.
Owner:TWIST BIOSCI
Construction method and application of artificial gene cluster plasmids containing pleocidin and multi-operon
The invention relates to a construction method and application of artificial gene cluster plasmids containing pleocidin and multi-operon. According to the construction method, efficient heterologous expression of macrolide bio-pesticide pleocidin can be realized by utilizing the DNA assembly technology and overexpression strategy driven by constructive promoters; 23 genes in the pleocidin synthesizing process are divided into five groups according to functions, and are respectively controlled by different constructive strong promoters to construct an artificial gene cluster of 102.6kb; and the23 genes bio-synthesized by pleocidin are divided into 7 operons, and the pleocidin yield of the artificial gene cluster in a heterologous host streptomyces albus J1074 is increased by 328 times in comparison with that of an original gene cluster. The multi-operon artificial gene cluster strategy provides a new method for synthetic biological researches of secondary metabolites.
Owner:SHANDONG UNIV
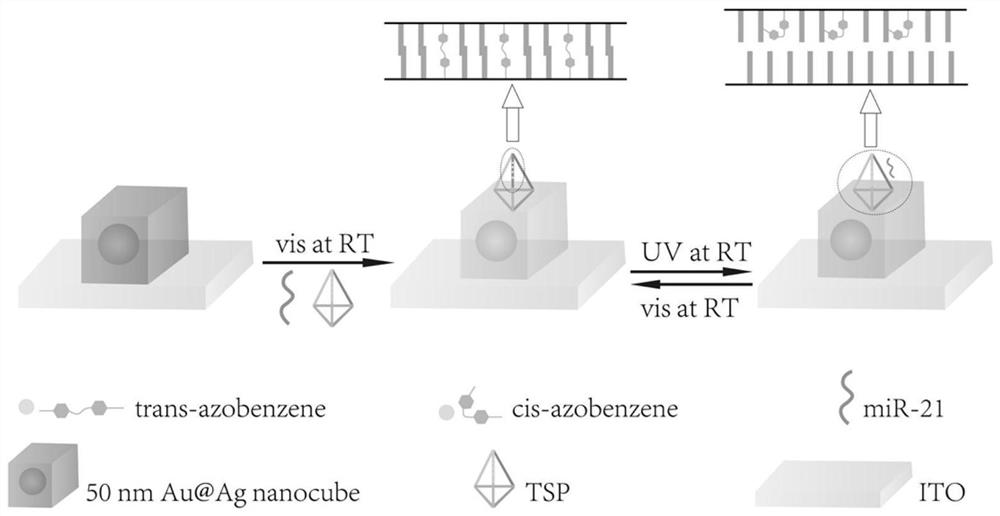
Biosensor as well as preparation method and application
ActiveCN111812064AAchieving High Sensitivity DetectionHigh detection sensitivityPreparing sample for investigationScattering properties measurementsNanoparticleDna assembly
The invention discloses a biosensor as well as a preparation method and application thereof. The biosensor disclosed by the invention is an assembly comprising gold and silver core-shell nanoparticle-tetrahedral structure DNA containing azobenzene molecules. The preparation method of the biosensor comprises the following steps of: preparing the gold and silver core-shell nanoparticles; preparing tetrahedral structure DNA containing the azobenzene molecules; and (3) assembling the gold and silver core-shell nanoparticle-tetrahedron DNA assembly containing the azobenzene molecules. According tothe biosensor which is the assembly comprising the gold and silver core-shell nanoparticle-tetrahedral structure DNA containing azobenzene molecules, high-sensitivity detection of miR-21 can be achieved on the single-nanoparticle scale, the concentration of miR-21 and the red shift amount of a scattering spectrum of the gold-silver core-shell nanoparticle-tetrahedral structure DNA assembly containing the azobenzene molecules are in a linear relation, and important significance is achieved for diagnosis and prognosis treatment of early-stage cancer.
Owner:NANJING UNIV OF POSTS & TELECOMM
Method of detecting target sarcosine of prostatic cancer
InactiveCN107760759AHigh detection sensitivityImprove the defect of poor assembly efficiencyMicrobiological testing/measurementDisease diagnosisDNA origamiProstate cancer
The invention provides a method of detecting target sarcosine of prostatic cancer. The method comprises the following steps: assembling SOx and HRP with DAN separately; synthesizing tubular DNA fold paper; assembling an enzyme-DNA compound with two types of DNA fold paper separately; assembling an SOx-origami compound III and an HRP-origami compound IV and detecting sarcosine; separately connecting cascaded enzymes Sox and HRP to the two types of tubular DNA fold paper; and achieving orderly cascading between the two enzymes by means of interaction of strong basic groups of complementary arm chains extending from the two types of fold paper, thereby achieving a purpose of improving detecting sensitivity. The novel detection method of regulating the distance between SOx and HRP orderly is established by means of the tubular fold paper structure. By way of further assembly after assembly of the enzymes and fold paper separately, the defect of poor assembling efficiency as a result of steric-hinderance effect is improved, and the assembling efficiency of the two enzymes is improved.
Owner:SHANGHAI NAT ENG RES CENT FORNANOTECH
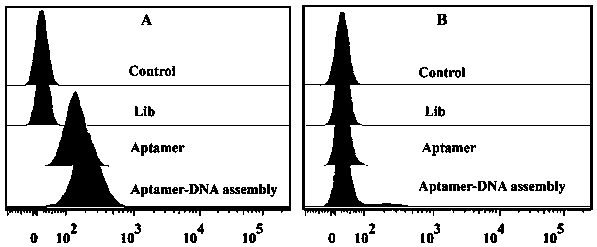
Construction method and application of aptamer-DNA macromolecular polymer based on non-linear hybridization chain amplification
ActiveCN109295133AAchieve therapeutic effectAchieve specific targetingOrganic active ingredientsPharmaceutical non-active ingredientsNucleic Acid ProbesA-DNA
The invention discloses construction and application of a DNA macromolecular polymer based on non-linear hybridization chain amplification. An initiating chain is introduced in the system, and six different non-hairpin nucleic acid probes are induced to perform cascading self-assembly to form the DNA macromolecular polymer through a non-linear hybridization chain reaction. On the basis, phosphorylase is used for treating a 5' end of a part of assembly sequence, and a DNA assembly with higher stability is formed under the action of ligase. Nucleic acid aptamers are combined at a viscous tail end to perform targeted recognition of target cells and are introduced into a cytotoxic drug doxorubicin, and imaging and drug administration of the target cells are achieved.
Owner:FUZHOU UNIV
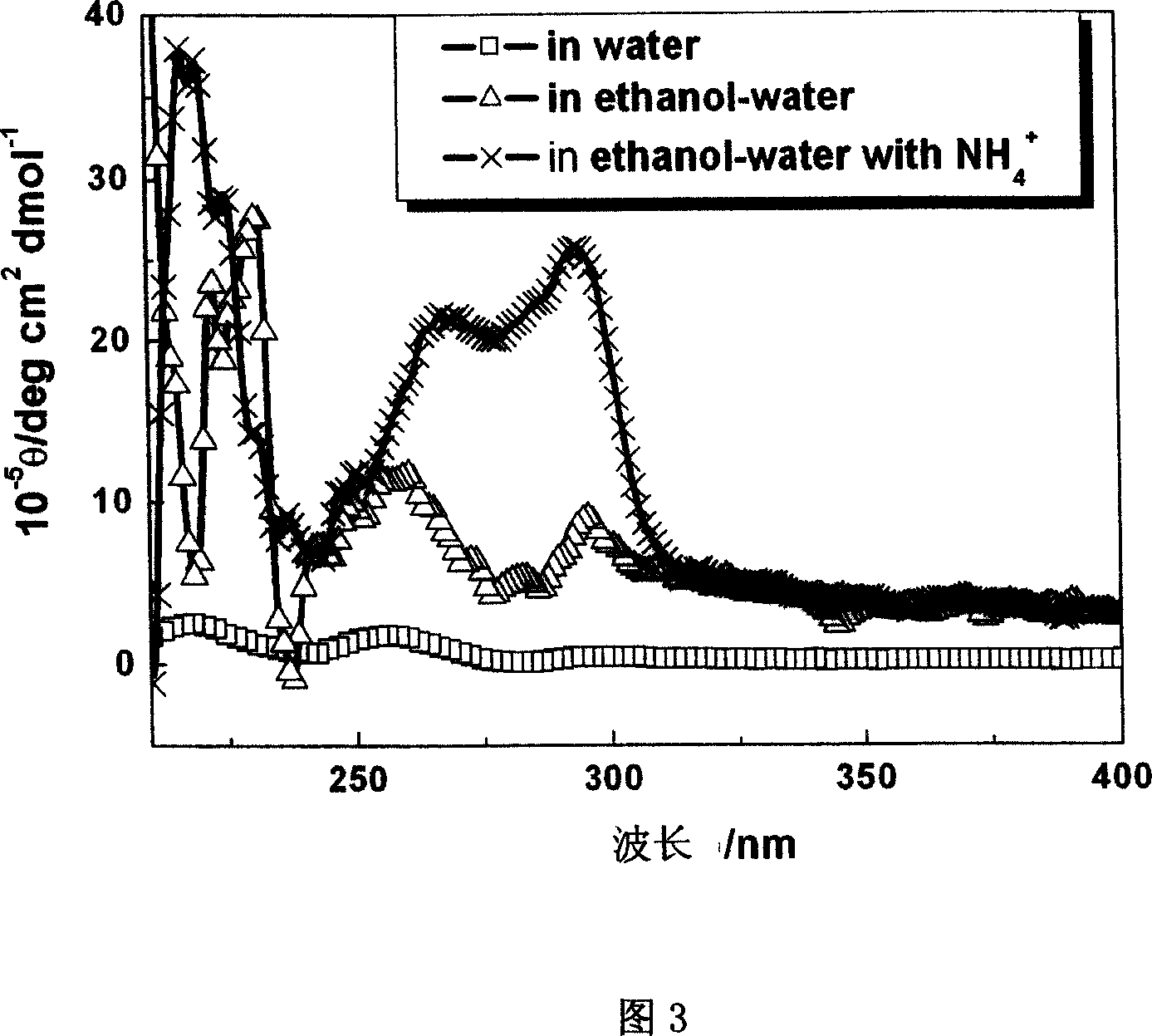
Micro array preparing DNA assembling process
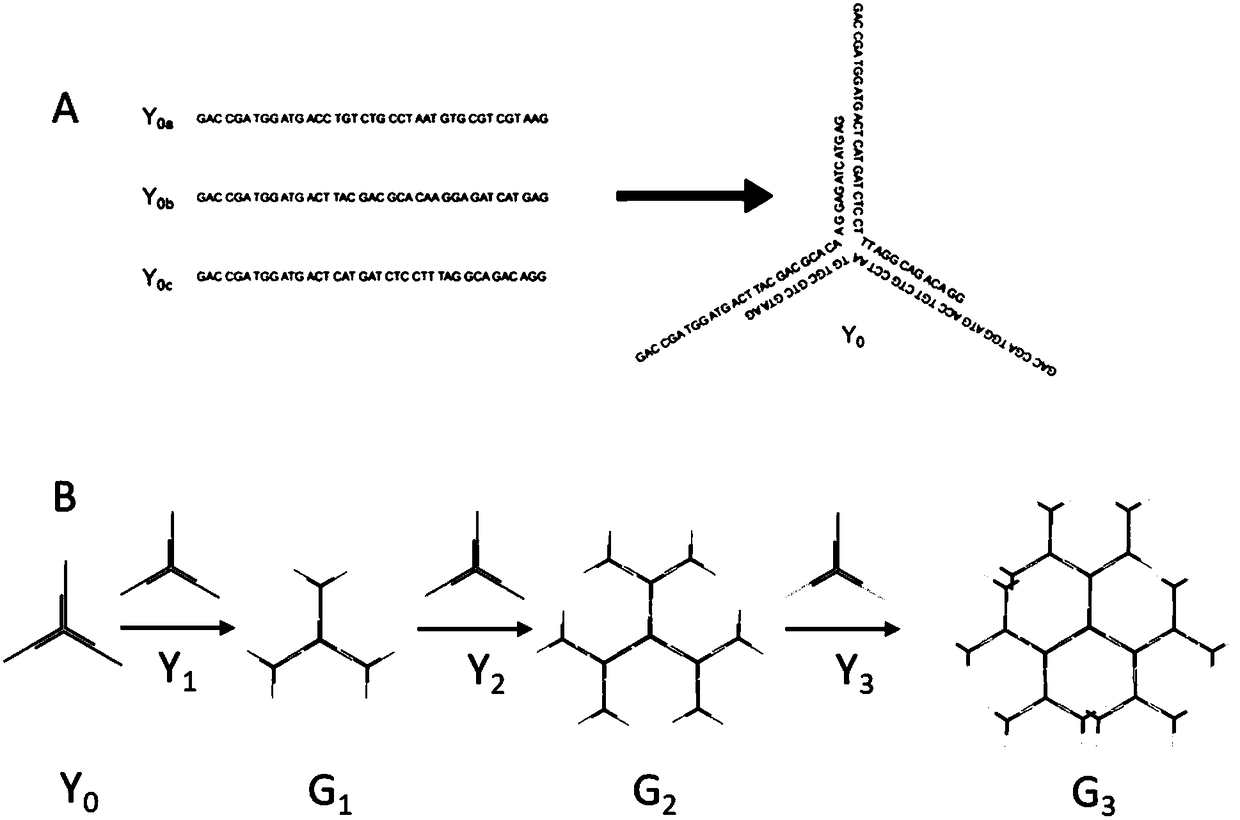
InactiveCN1986831ARegulate branching structureEasy to manufactureMicrobiological testing/measurementAlcoholMetal particle
The micro array preparing DNA assembling process includes the following steps: 1. dissolving DNA sequence in pure water, mixing with low molecular weight alcohol or ketone, and adding water solution containing Li+, NH4+, Na+ or K+; 2. heating to 80-95 deg.c, cooling to 20-30 deg.c, and incubating at 2-25 deg.c for 10-24 hr; and 3. soaking mica substrate in the solution obtained in the last step for 10-50 min, taking out and drying to obtain DNA assembling micro array. The preparation process is simple, and the prepared DNA assembling micro array has branched structure capable of being regulated through selecting DNA sequence and solution metal particle. The present invention can promote the development of DNA molecular device, DNA circuit, micro channel manufacture, etc. and the DNA assembling micro array may be used as new regular template for guiding the preparation of functional device or material.
Owner:TIANJIN UNIV
Novel oligo-linker-mediated DNA assembly method and applications thereof
InactiveUS20160340670A1Efficient assemblyAvoid introducingNucleic acid vectorVector-based foreign material introductionBiological pathwayOligonucleotide Linker
A method for generating a library of expression vectors comprising a plurality of donor sequences and a plurality of oligo-linker nucleic acids, termed Oligonucleotide Linker-Mediated DNA Assembly (OLMA), is described. Also described are applications of the OLMA method, including the simultaneous tuning of several factors in metabolic and biological pathways, and the combinatorial high throughput optimization of metabolic and biological pathways.
Owner:NANJING GENSCRIPT BIOTECH CO LTD
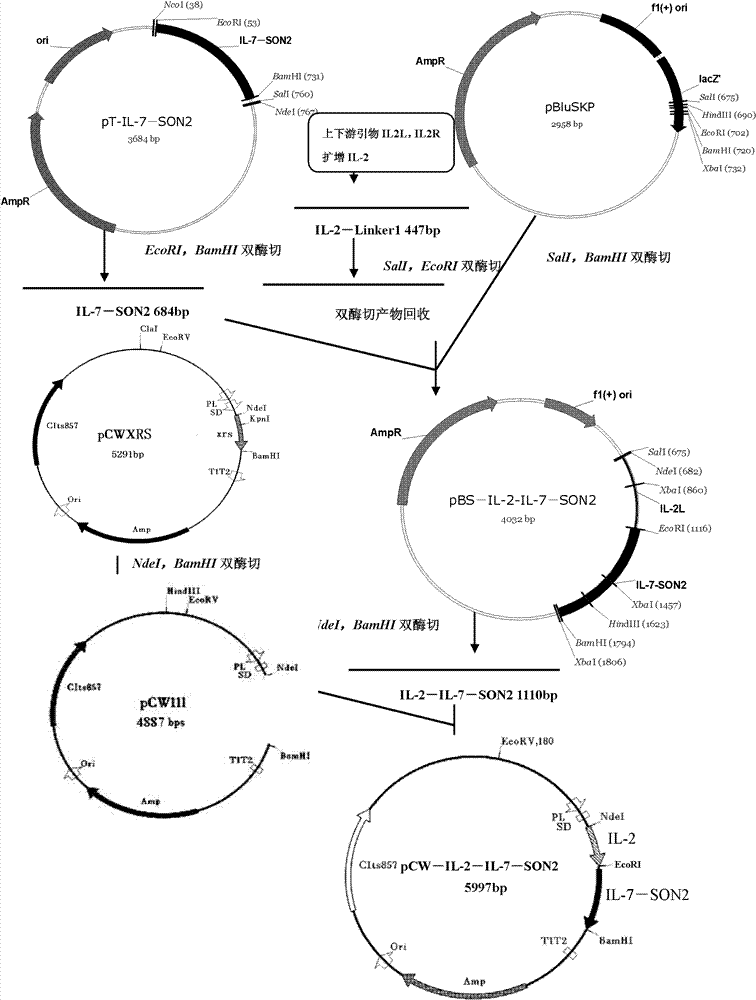
Composite medicine for targeted elimination of HIV/SIV
InactiveCN102775496AInhibition of reproductionAccurate and strict controlGenetic material ingredientsAntiviralsDna assemblyViral vector
The invention relates to composite medicine for targeted elimination of HIV / SIV. Specifically, the invention relates to a fusion protein, which is composed of human cytokine polypeptide IL2, IL7, DNA binding protein SON2 rich in positive amino acids, as well as a co-receptor polypeptide CXCR4 able to achieve specific binding with gp120 and / or gp41 and neutralization of a free HIV virus, and serves as a non-viral vector. The non-viral vector fusion protein and a restrictive toxin gene expression recombinant DNA can be assembled to form the composite medicine. The invention also relates to application of the fusion protein and the composite medicine in elimination of HIV in AIDS (acquired immune deficiency syndrome) patients.
Owner:THE INST OF BASIC MEDICAL SCI OF CHINESE ACAD OF MEDICAL SCI +1
DNA assembly and cloning method
ActiveCN104357438AShort reaction timeReduce the chance of mutationDNA preparationDna assemblyPolygene
The invention belongs to the field of biotechnology and discloses a simple DNA assembly and cloning method. According to the DNA assembly and cloning method, a polymerase chain reaction between an insert fragment and a linearized vector which are used as a primer and a template mutually is carried out to obtain double-stranded circular DNA, wherein two ends of the insert fragment respectively carry overlapping sequences overlapping with two ends of the linearized vector, and two ends of the linearized vector respectively carry overlapping sequences overlapping with two ends of the insert fragment. The DNA assembly and cloning method has a simple reaction system, is low-cost, is simple to operate, is easy for parallel operation, has obvious advantages for DNA assembly and cloning of a complex gene fragment library, a combined gene fragment library and polygene pathways, can widely be applied in DNA assembly and cloning of a single gene, polygenes and a gene fragment library, and is especially suitable for high-throughput gene synthesis and assembly.
Owner:TIANJIN INST OF IND BIOTECH CHINESE ACADEMY OF SCI
Seamless DNA assembling method being high in efficiency and low in cost
InactiveCN110592130ALow costShorten the durationVector-based foreign material introductionEscherichia coliDNA fragmentation
A method provided by the invention comprises the steps of performing engineering reforming on escherichia coli DH10B, knocking out recB-recC-rec-D genes on a genome, and performing overexpression on recE-recT genes on the genome to obtain a recombinant engineering host bacterial strain DH10B* being good in performance; and co-transferring 1-3 DNA fragments with 20bp-40bp homologous sequence jointsinto cells to realize in vivo assembly of a complete plasmid, wherein the positive rate of colonies formed on a flat is higher than 95%. Compared with a conventional seamless DNA assembling method, the method disclosed by the invention omits expensive and time-consuming in vitro connection operation, realizes high-efficiency, low-cost and low-time-consumption seamless DNA assembling, and has great application value in molecular biology experiments.
Owner:BEIJING INSTITUTE OF TECHNOLOGYGY
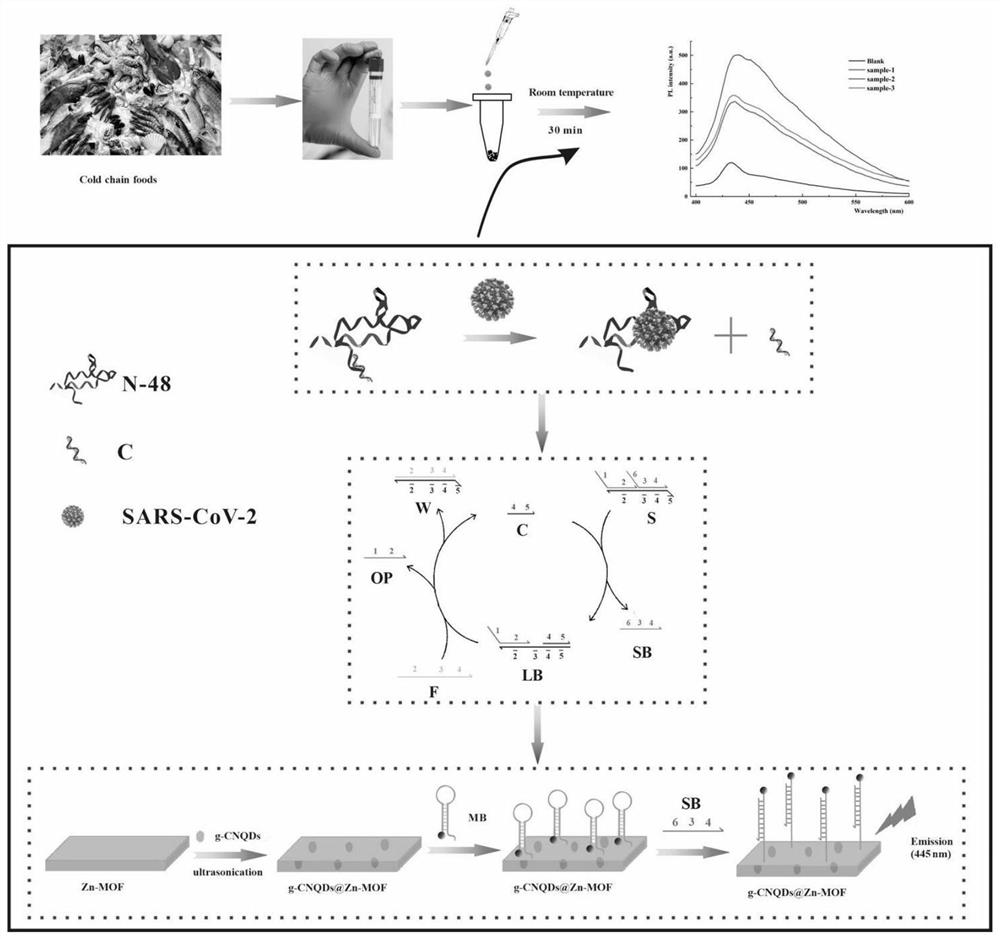
Construction method of fluorescent aptamer sensor and application of fluorescent aptamer sensor in novel coronavirus detection
ActiveCN114002425AImprove stabilityGood repeatabilityFluorescence/phosphorescenceImmunoassaysConformational entropyDna assembly
The invention discloses a preparation method of a fluorescent aptamer sensor based on conformation entropy driving DNA assembly reaction. The preparation method comprises the following steps: (1) preparing g-CNQDs; (2) preparing Zn-MOF; (3) preparing a g-CNQDs@Zn-MOF solution: synthesizing the g-CNQDs@Zn-MOF solution from the Zn-MOF and the g-CNQDs; (4) carrying out biological functionalization on the g-CNQDs@Zn-MOF: connecting an MB chain to the g-CNQDs@Zn-MOF; and (5) forming the fluorescent aptamer sensor. The sensor is used for detecting the novel coronavirus, and analysis can be completed within 30 min.
Owner:SICHUAN UNIV
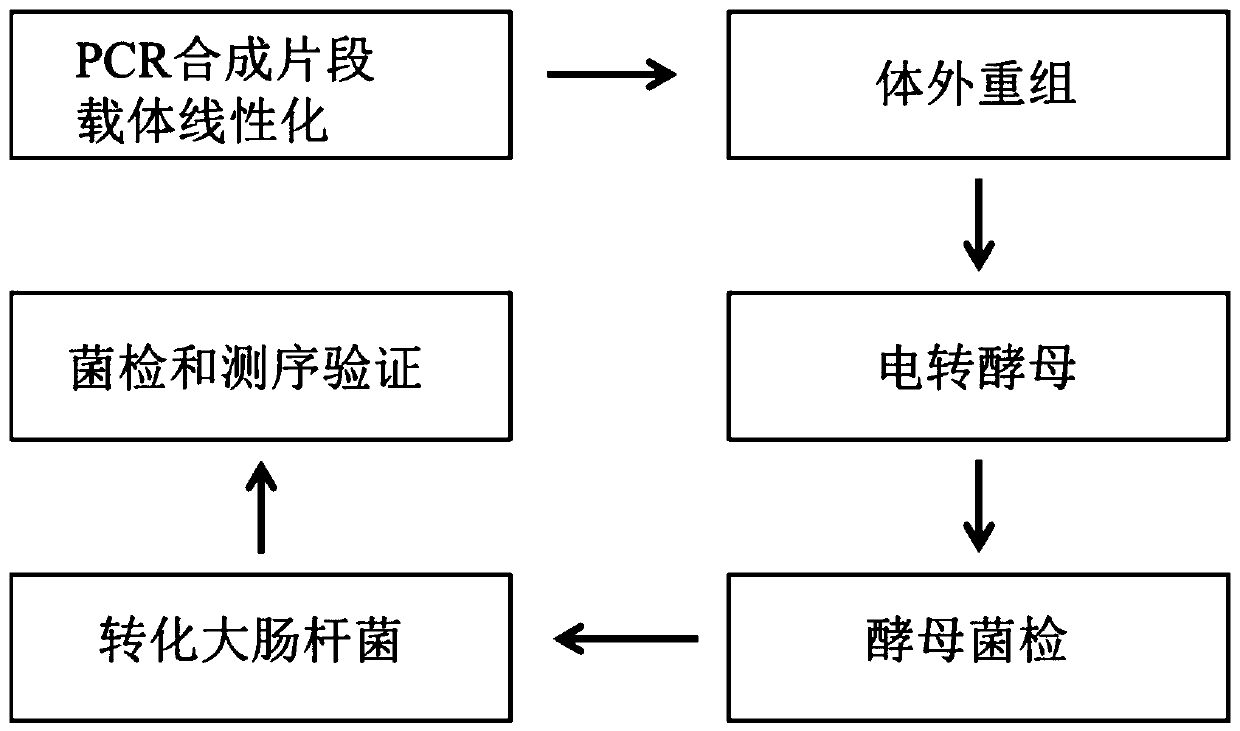
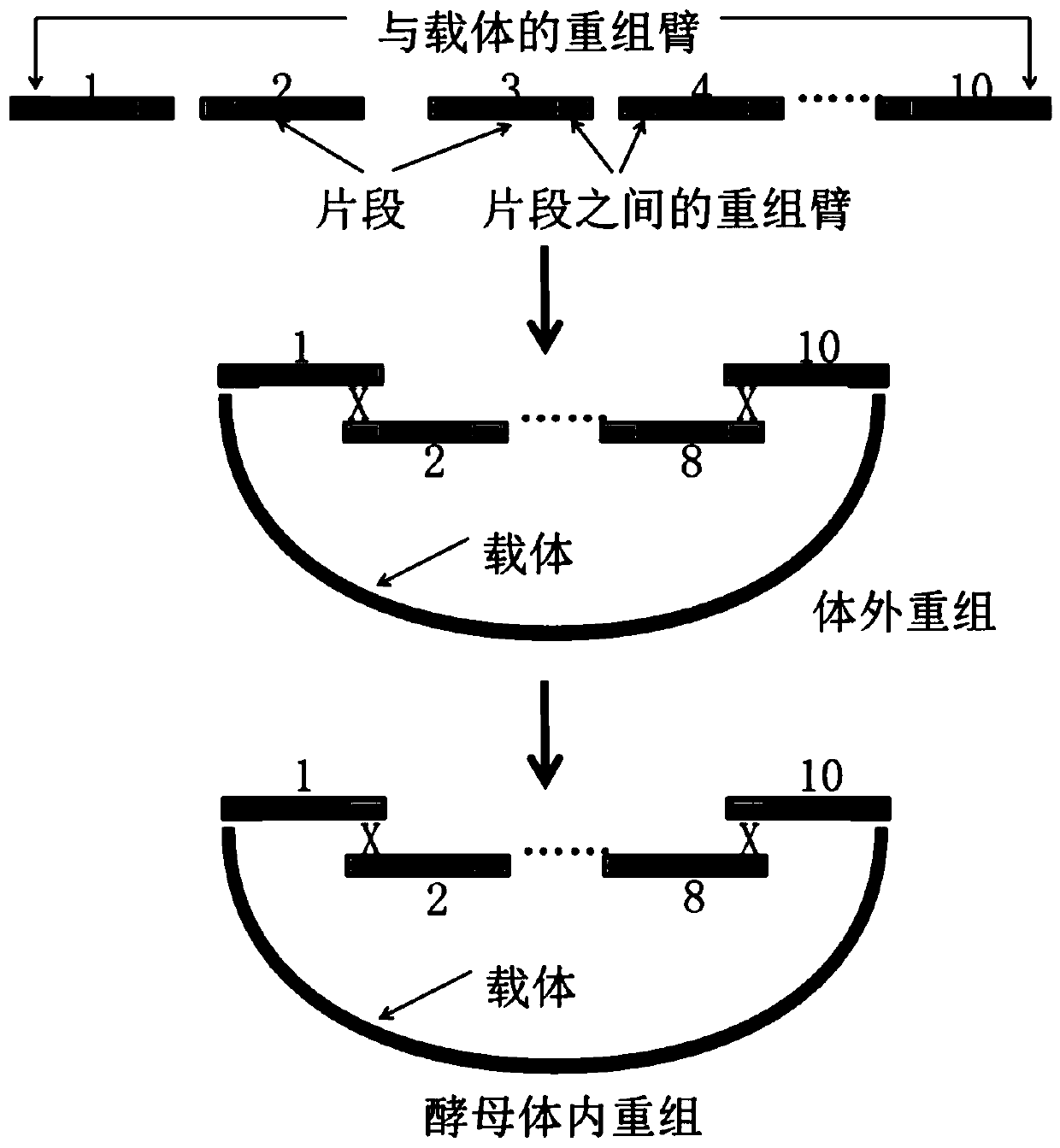
Multi-fragment DNA assembly kit, multi-fragment DNA assembly method and application thereof
PendingCN110964738ALower success rateIncrease the lengthMicroorganism based processesVector-based foreign material introductionDna assemblyRecombinase
The invention provides a multi-fragment DNA assembly kit, a multi-fragment DNA assembly method and application thereof. The kit comprises a to-be-assembled DNA fragment Fn and a vector. The vector includes an intermediate connection vector and / or an assembly vector; the kit further comprises saccharomyces cerevisiae. According to the invention, a mode of combining in-vitro assembly and in-vivo assembly is adopted; an initial fragment is synthesized through PCR and the initial fragment is added into the intermediate connection vector, a recombinant enzyme system is adopted to assemble large-fragment DNA and the assembly vector in vitro, and then an in-vitro assembly product is converted into saccharomyces cerevisiae for in-vivo assembly. Advantages of in-vitro assembly and in-vivo assemblyare combined and the synthesis success rate of super-large plasmids is remarkably increased.
Owner:SUZHOU HONGXUN BIOTECH CO LTD
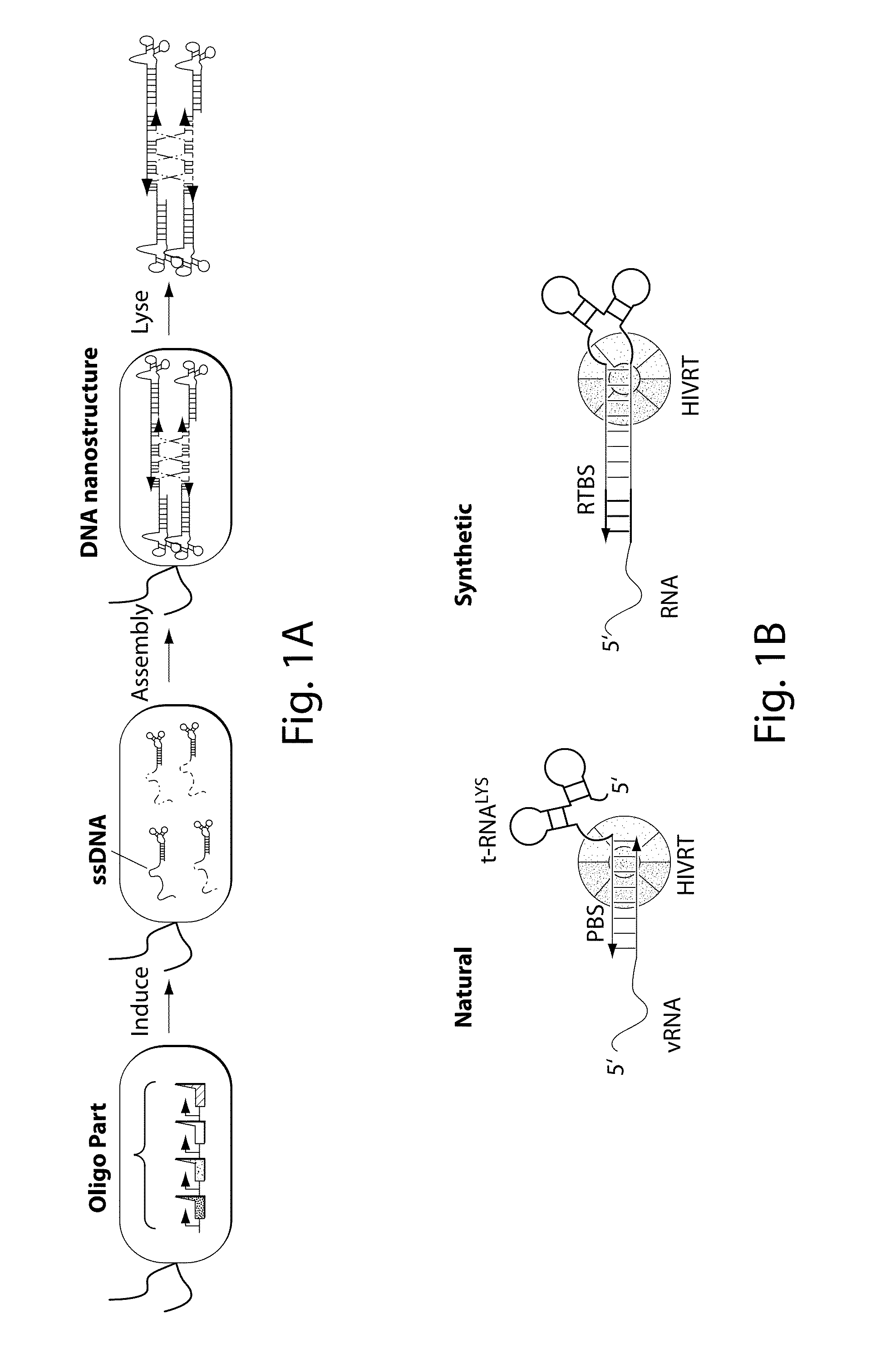
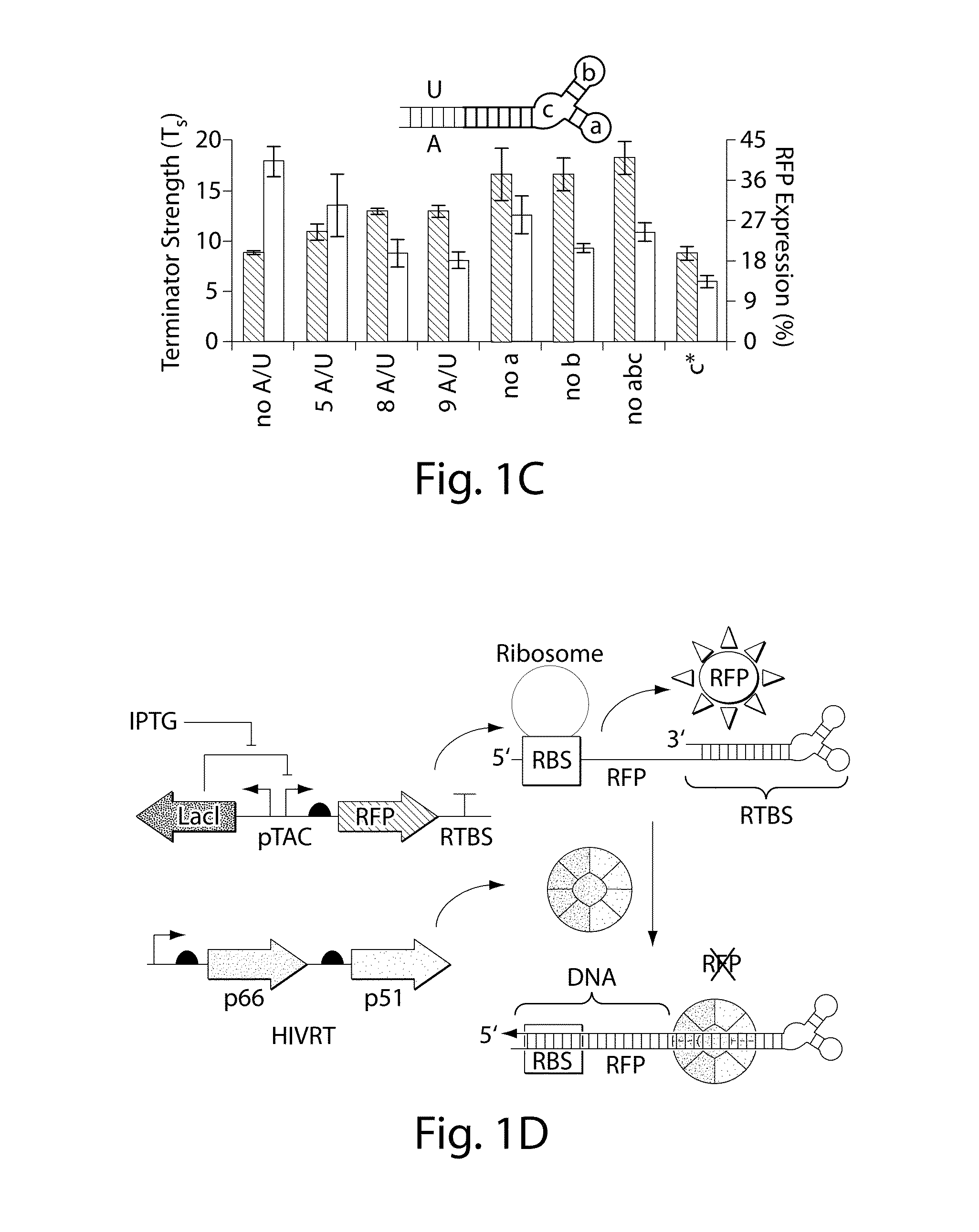
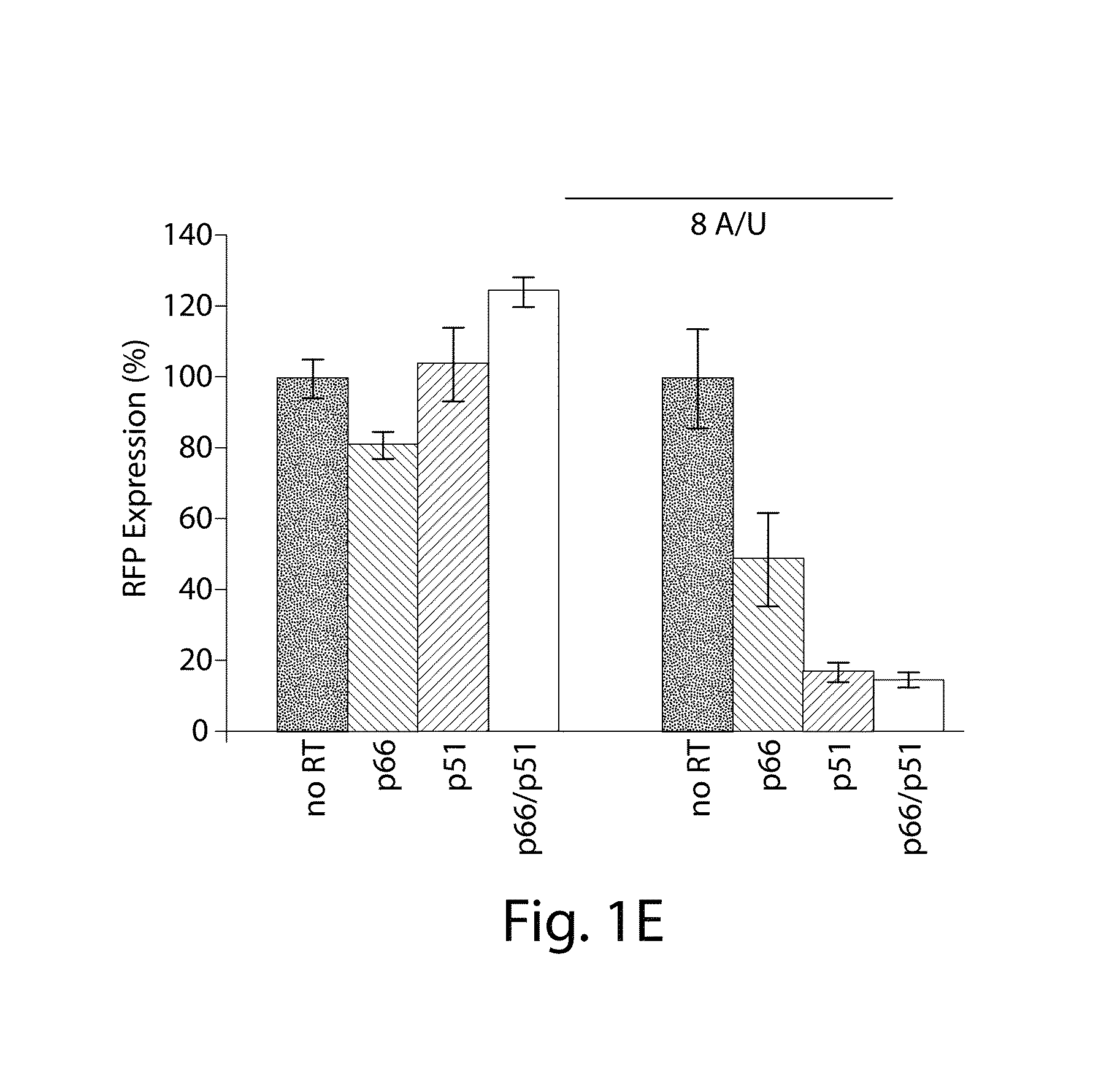
Engineering DNA assembly in vivo and methods of making and using the reverse transcriptase technology
Cells that can synthesize oligonucleotides in vivo to produce a nucleic acid nanostructure are described. Methods for producing oligonucleotide nanostructures for use in regulating gene expression and altering biological pathways are provided. Methods of performing multiplex automated genome editing (MAGE) are also provided.
Owner:MASSACHUSETTS INST OF TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com