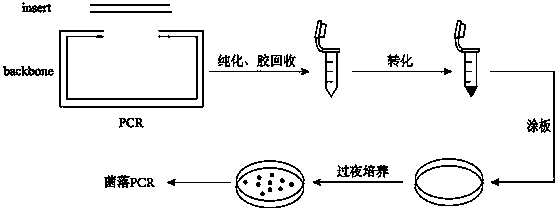
Seamless DNA assembling method being high in efficiency and low in cost
A low-cost, high-efficiency technology, applied in the field of bioengineering, can solve problems such as cumbersome operation procedures and expensive kits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0024] (1) Production of DH10B*transformation competent cells: ①First pick a single colony from the DH10B* plate, put it into 5ml of liquid LB medium and culture it overnight in a shaker at 37°C; ②From the culture Take 50μl from the solution and add it to a new 50ml liquid medium and culture it at 37℃ to OD 600 When it reaches 0.3, add 80 μl of IPTG with a concentration of 100 mM, and then put it in a shaker to continue culturing until OD 600 reach 0.5-0.6; ③ Shake the 50ml bacterial solution, put it in an ice bath for 30 minutes, and turn on the centrifuge at the same time and adjust it to 4°C for pre-cooling; ④ After 30 minutes (all operations are carried out at 4°C), transfer the bacterial solution into the centrifuge in the ultra-clean bench tube, and centrifuge at 5000rpm at 4°C for 2min to precipitate the bacteria; ⑤Pour off the supernatant and add 22.5ml 0.1M Cacl 2 After resuspension, let stand for 5-10min, then centrifuge at 5000rpm, 4°C for 2min to precipitate the b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com