Colorimetric method for simultaneously detecting multiple miRNA (micro-ribonucleic acid) sequences based on competitive hybridization reaction
A technology of hybridization reaction and DNA sequence, which is applied in biochemical equipment and methods, measuring devices, microbe determination/inspection, etc., and can solve problems such as low sensitivity, inability to distinguish homologous miRNA sequences, and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Effect of hybridization temperature (determination of miRNA-182 sequence):
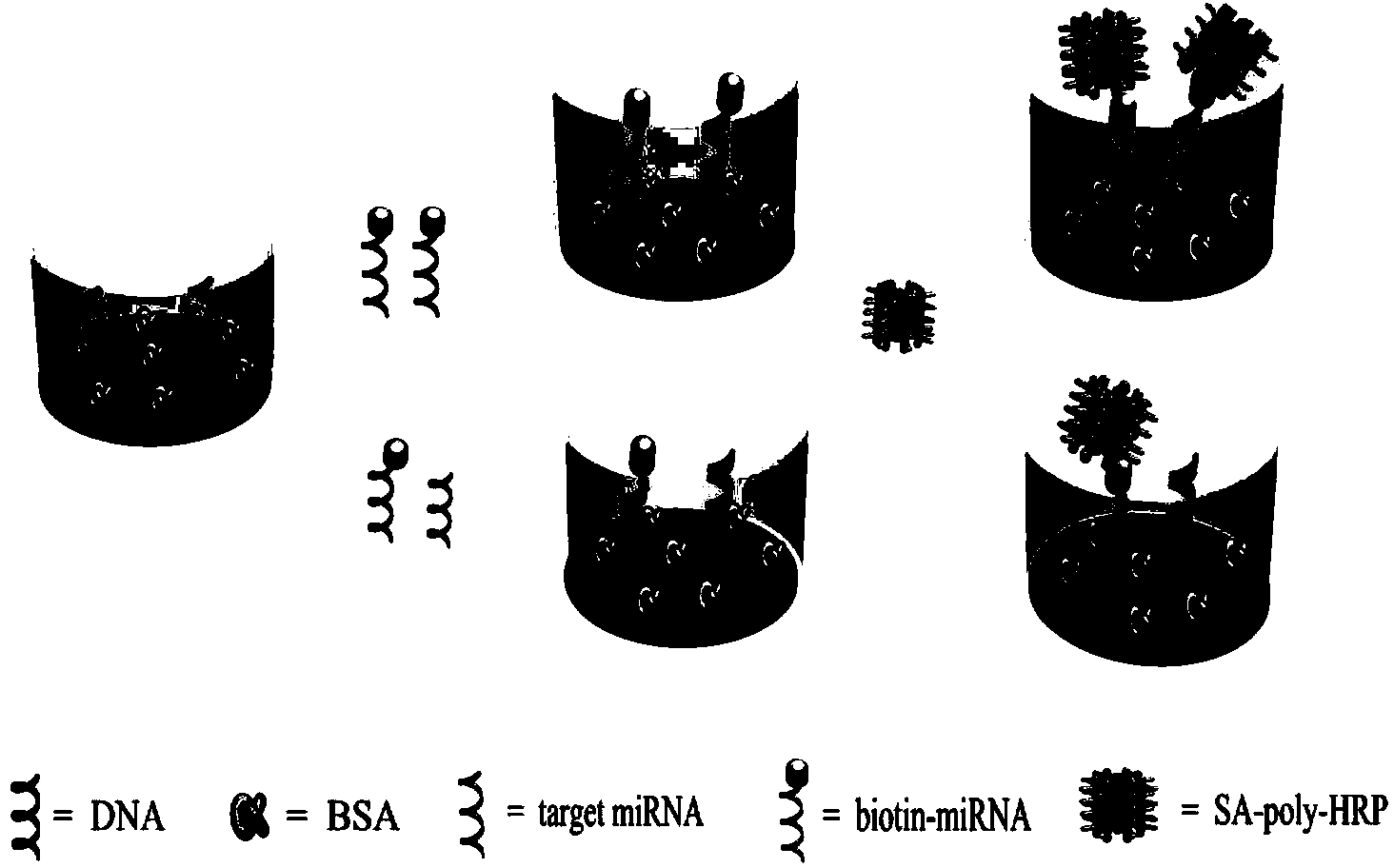
[0059] (1) Add 60 μL of 1 nM amino DNA solution (sequence 3'-AAA CCG TTA CCA TCT TGA GTG TGA TTT TT-(CH 2 ) 6 NH 2 -5'), let stand overnight, and wash with PBS buffer several times;
[0060] (2) Block with 180 μL of 2% BSA solution at room temperature for 2.5 hours, and then wash with PBS buffer several times;
[0061] (3) On the basis of the above step (2), add 120 μL of biotin-labeled miRNA-182 solution (sequence 3'-UCA CAC UCA AGA UGG UAA CGG UUU-biotin-5') with a concentration of 1 nM dropwise, respectively Placed at 25°C, 30°C, 37°C, 45°C, 50°C, reacted for 4 hours, washed with PBS buffer several times;
[0062] (4) On the basis of the above step (3), add 60 μL SA-Poly-HRP solution dropwise, and after standing at room temperature for 1 hour, wash with PBS buffer several times;
[0063] (5) Mix 100 μL of 0.1% TMB solution and 25 μL of 0.03% H 2 o 2 The solution was successively added...
Embodiment 2
[0080] The influence of hybridization temperature and hybridization time is as embodiment 1;
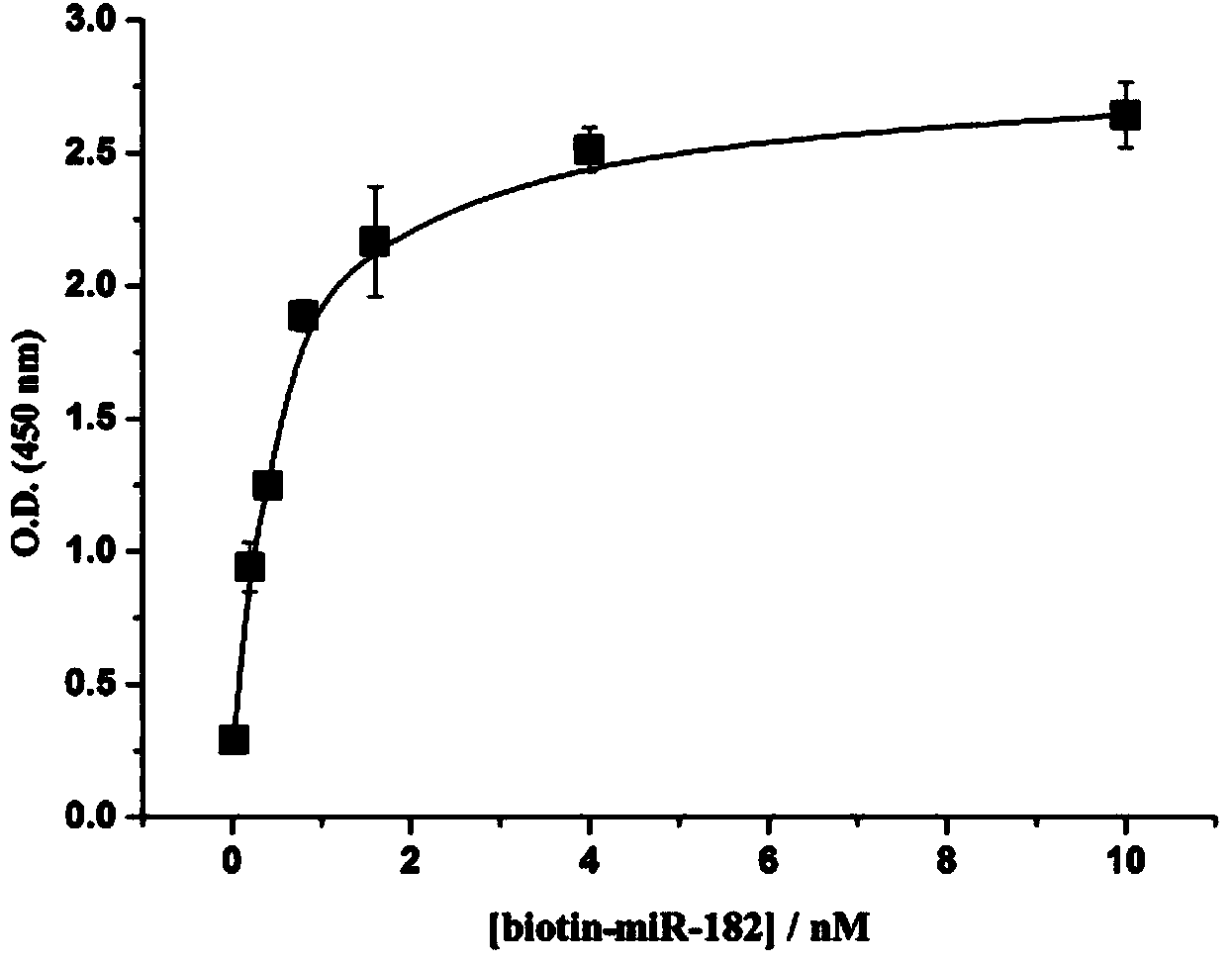
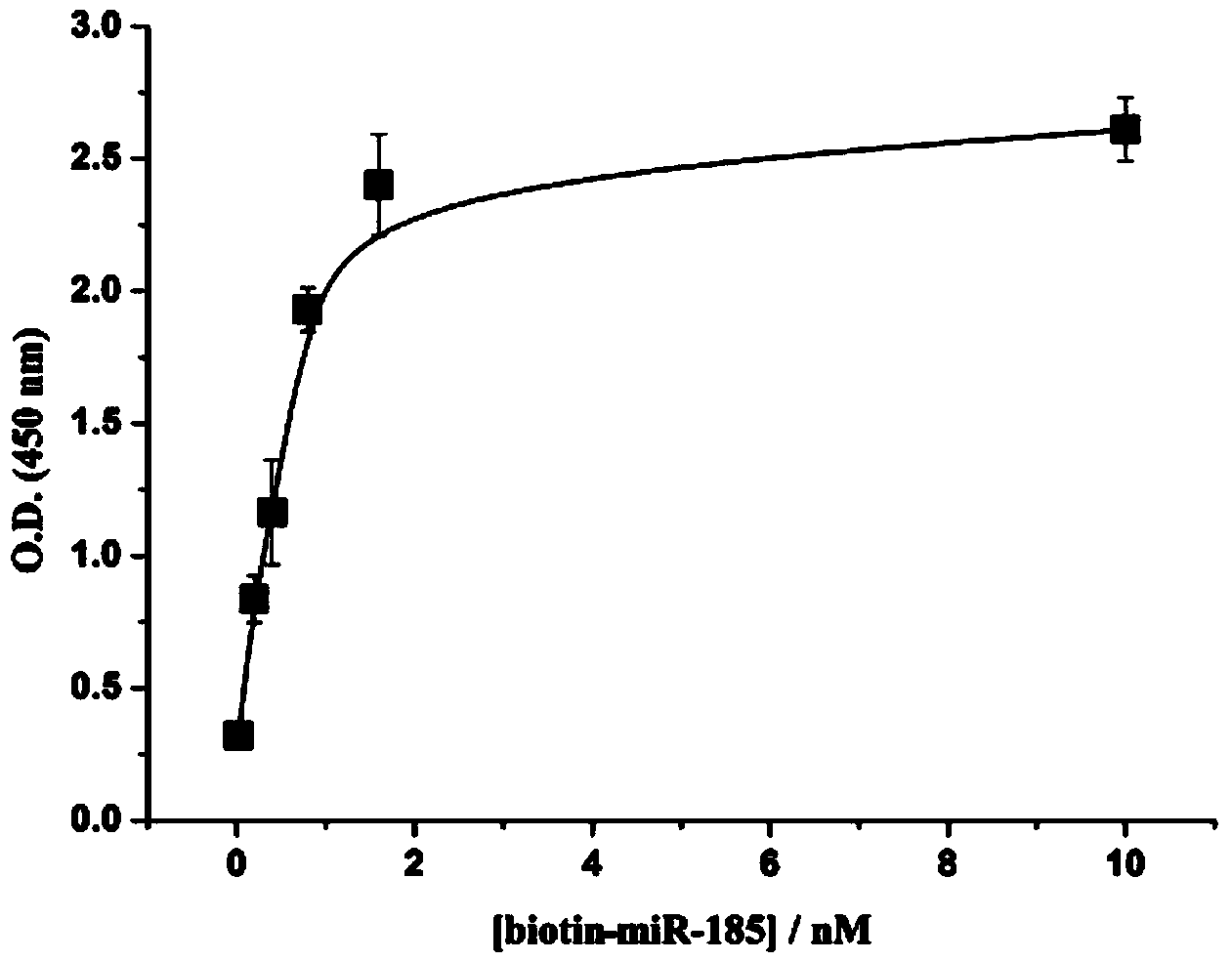
[0081] The content detection experiment of miRNA-185 (sequence 5'-UGG AGA GAA AGG CAG UUC CUGA-3') in serum:
[0082] Effect of different concentrations of biotin-labeled miRNA sequence (identical to miRNA-185 sequence):
[0083] (1) Add 60 μL of 1 nM amino DNA solution dropwise to the microtiter plate, let it stand overnight, and wash it with PBS buffer several times;
[0084] (2) Block with 180 μL of 2% BSA solution at room temperature for 2.5 hours, and then wash with PBS buffer several times;
[0085] (3) On the basis of step (2), drop a series of 120 μL biotin-miRNA solutions with different concentrations in the microwells, the concentrations are 0, 0.1, 0.2, 0.5, 0.8, 1.0, 2.0, 5.0, 10 nM , placed at 37°C for 2.5 hours, washed with PBS buffer several times;
[0086] (4) On the basis of step (3), add 60 μL of SA-Poly-HRP solution dropwise into the microwells, let stand at roo...
Embodiment 3
[0092] The influence of hybridization temperature and hybridization time is as embodiment 1;
[0093] The content detection experiment of miRNA-381 (sequence is 5'-UAU ACA AGG GCA AGC UCU CUGU-3') in serum:
[0094] The effect of different concentrations of biotin-labeled miRNA sequence (same sequence as miRNA-381):
[0095] (1) Add 60 μL of 10 nM amino DNA solution dropwise to the microtiter plate, let it stand overnight, and wash it with PBS buffer several times;
[0096] (2) Block with 180 μL of 2% BSA solution at room temperature for 2.5 hours, and then wash with PBS buffer several times;
[0097] (3) On the basis of step (2), drop a series of 120 μL biotin-miRNA solutions with different concentrations in the microwells, the concentrations are 0, 0.1, 0.2, 0.5, 0.8, 1.0, 2.0, 5.0, 10 nM , placed at 37°C for 2.5 hours, then rinsed with PBS buffer several times;
[0098] (4) On the basis of step (3), add 60 μL of SA-Poly-HRP solution dropwise into the microwells, let stan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com