Nuclease-mediated DNA assembly
A technology of nuclease and exonuclease, which is applied in the direction of DNA preparation, recombinant DNA technology, and the introduction of foreign genetic material using vectors, etc. It can solve problems such as limited accuracy, time-consuming multi-step reactions, and unpredictable off-target effects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0178] The following examples are given to provide those of ordinary skill in the art with a complete disclosure and description of how to make and use the invention, and are not intended to limit the scope of what the inventors regard as their invention, nor are they intended to represent that the following experiments are All or only experiments performed. Efforts have been made to ensure accuracy with respect to numbers used (eg amounts, temperature, etc.) but some experimental errors and deviations should be accounted for. Unless indicated otherwise, parts are parts by weight, molecular weight is weight average molecular weight, temperature is in degrees Celsius, and pressure is at or near atmospheric.
example 1
[0179] Example 1: BAC digestion with CAS9 and assembly with selection cassette
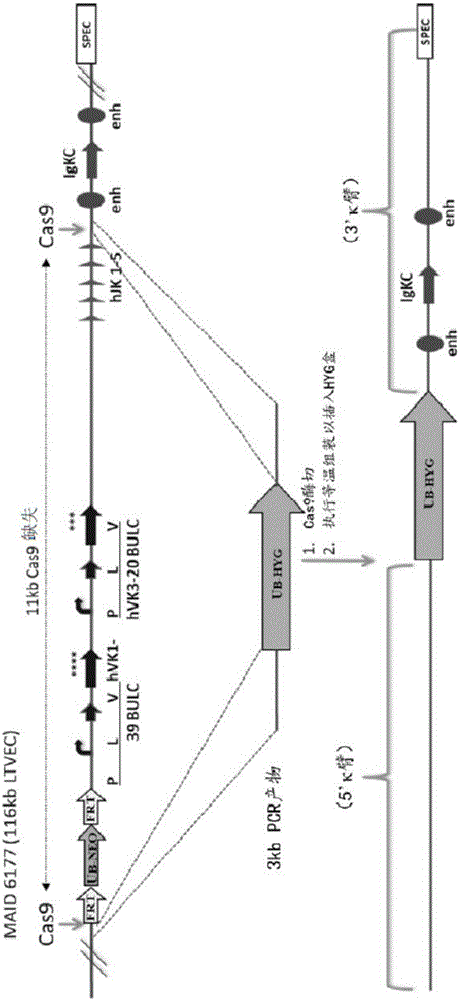
[0180] Artificial crRNA and artificial tracrRNA were designed to target specific sequences in MAID 6177 (116 kb LTVEC) for assembly with a 3 kb PCR product (UB-HYG). The PCR product contained a 50 bp region overlapping the vector. crRNA and tracrRNA were first dissolved in duplex buffer (30 mM HEPES, pH 7.5, 100 mM potassium acetate) to 100 μM. To anneal the RNA, add 10 µL of 100 µM crRNA and 10 µL of 100 µM tracrRNA to 80 µL of annealing buffer. Heat the RNA in a 90°C heating block, then remove the heating block from the heater and allow to cool on the bench. The final concentration of RNA was about 10 μM.
[0181] To digest BAC, use clean maxiprep BAC DNA and digest BAC according to the mixture below.
[0182]
[0183]
[0184] Digest at 37°C for 1 hour, then desalt for 30 minutes. The final reaction buffer contained: 20 mM Tris 7.5; 100-150 mM NaCl; 10 mM MgCl2; 1 mM DTT; 0.1 mM ED...
example 2
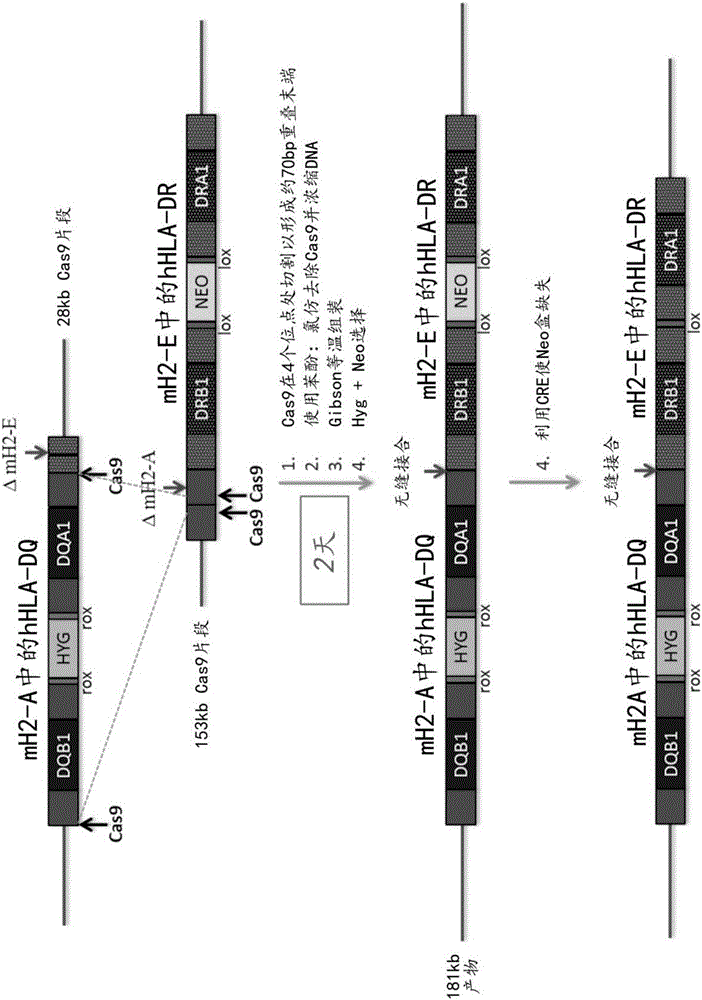
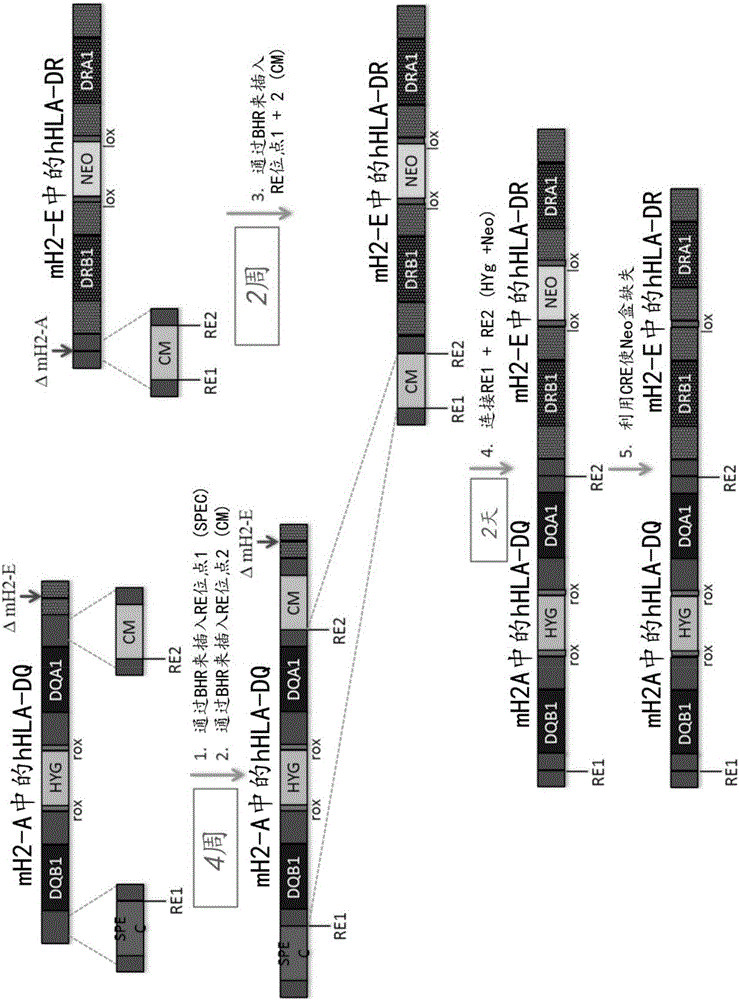
[0194] Example 2: Stitching together two overlapping BACs: humanization in the mouse MHC II locus (H2-A / H2-E) HLA-DQ+Humanized HLA-DR
[0195] Artificial crRNA and artificial tracrRNA were designed to target specific sequences in humanized HLA-DQ BAC for assembly with humanized HLA-DR BAC. These vectors contain approximately 70 bp overlapping regions formed by Cas9 cleavage at two sites on each vector (see figure 2 ). Dissolve crRNA and tracrRNA in Hybe buffer to 100 μM. To anneal the RNA, add 10 µL of 100 µM crRNA and 10 µL of 100 µM tracrRNA to 80 µL of annealing buffer. Place the RNA in a 90°C heating block, then remove the heating block from the heater and allow to cool on the bench. The final concentration of RNA was about 10 μM.
[0196] For digestion of BAC, clean maxiprep BAC DNA can be used. Digest each BAC individually according to the following mix:
[0197]
[0198] The BAC vector should be digested at 37°C for 1 hour and then heat inactivated at 65°C...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com