Multi-fragment DNA assembly kit, multi-fragment DNA assembly method and application thereof
A kit and multi-fragment technology, applied in the field of synthetic biology, can solve the problems of low assembly success rate, long time-consuming and high cost, and achieve the effect of high success rate, speeding up synthesis rate and reducing cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] In vitro assembly of embodiment 1 large fragment DNA
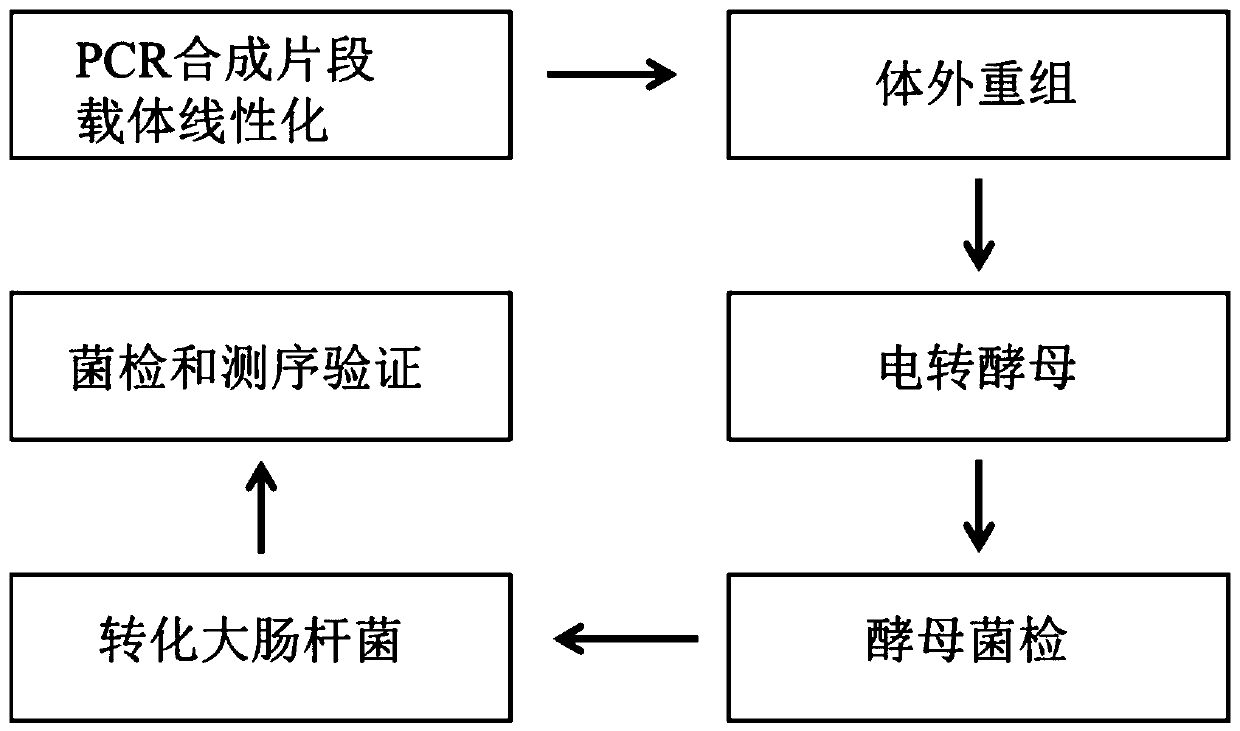
[0071] The construction process of large fragment recombinant plasmids is as follows: figure 1 As shown, in this example, the in vitro assembly of large fragments of DNA was first carried out.
[0072] (1) Splitting of large fragments of DNA
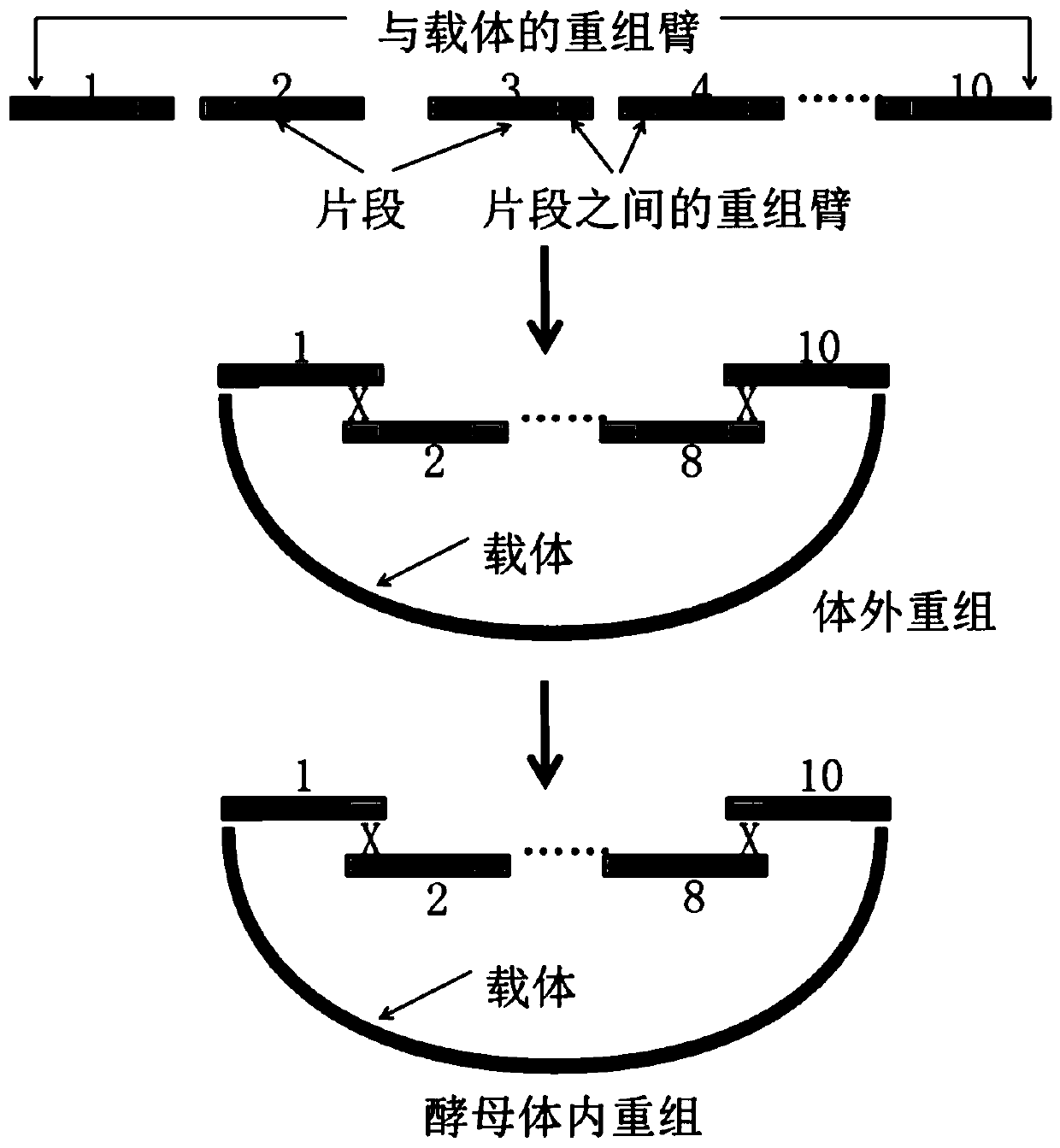
[0073] Use sequence analysis software to analyze large fragments of DNA with a length of 20 kb (such as image 3 ) for analysis, split into 10 initial fragments with a length of 2kb (such as image 3 ), and design homology arms between adjacent initial fragments (such as image 3 ) (homologous arms are the leftmost 60bp sequence and the rightmost 60bp sequence of the initial fragment), so that subsequent homologous recombination can be performed;
[0074] (2) Splitting of the initial segment
[0075] Use sequence analysis software to split the initial fragments into DNA fragments to be assembled F n , n=20, where, F n It is a long primer sequence separated by forward an...
Embodiment 2
[0089] Example 2 Preparation of Saccharomyces cerevisiae Electrotransfer Competent Cells
[0090] (1) Cultivate Saccharomyces cerevisiae overnight, reach the stable phase, and the OD600 reaches 3;
[0091] (2) Inoculate part of the yeast culture solution into 100mL YPD medium (10g / L yeast nitrogen base, 20g / L peptone, 20g / L D- -glucose), make the OD600 value reach 0.03, cultivate the yeast until the OD600 reaches 1.6, and collect the thalline by centrifugation;
[0092] (3) Wash the yeast pellet once with 50mL cold water, transfer it to a 20mL 0.1M LiAc / 10mM DTT Erlenmeyer flask, and treat it for 30min at 30°C and 225rpm on a shaking table;
[0093] (4) Wash the yeast pellet twice with 50 mL of cold water and once with 50 mL of 1M sorbitol;
[0094] (5) Suspend the yeast pellet in 100-200mL 1M sorbitol, then centrifuge and concentrate to 1mL, so that the concentration of yeast is equivalent to 1.6×10 9 Cells / mL, aliquoted for storage.
Embodiment 3
[0095] In vivo assembly of embodiment 3 large fragment DNA
[0096] Purify the in vitro assembly product obtained in Example 1 through a column, take 10 μL of the purified product and add it to Saccharomyces cerevisiae electroporation competent cells, mix well, set the electroporation program
[0097] The mixture of the purified large-fragment DNA and the assembled vector was electroporated into Saccharomyces cerevisiae competent cells by electroporation with a BioRad GenePulser cuvette (the distance between the electrodes is 0.2 cm), and the shock duration was 5 ms;
[0098] After electroporation, Saccharomyces cerevisiae cells were inoculated in 1 mL of sorbitol:YPD (1:1) medium with a concentration of 1 M, and cultured in suspension at 30°C for 1 h;
[0099] The collected cells were inoculated on the screening marker plate and cultured at 30°C for 3 days. The colony diagram is as follows: Figure 5 shown;
[0100]Pick a single colony with a sterilized toothpick and place ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com