A kind of circRNA, product and application thereof
A product and sample technology, applied in the field of agricultural genetic engineering, can solve problems such as fat buttocks, and achieve the effect of improving meatiness and meat performance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] The collecting agent RNA of embodiment 1 sample is extracted
[0041] 1.1 Collection of samples
[0042] In this experiment, Duolang sheep (D) and Small Tail Han sheep (X) with different fat deposits were studied. The selected sheep are all adult females, and the individuals are in good body condition, strong and disease-free, with similar body weight (about 50kg), and each group has three biological replicates.
[0043] 1.2 Extraction and quality detection of sample RNA
[0044] Take an equal amount of adipose tissue from each sample to extract total RNA, and ensure that the whole process is carried out in a sterile environment. The steps are as follows:
[0045] (1) Grinding adipose tissue in a mortar with liquid nitrogen;
[0046] (2) Put the ground tissue into a centrifuge tube added with Trizol reagent and let it stand;
[0047] (3) Add chloroform and mix well and let stand at room temperature for 10 minutes;
[0048] (4) Put it into a centrifuge, and centrifu...
Embodiment 2
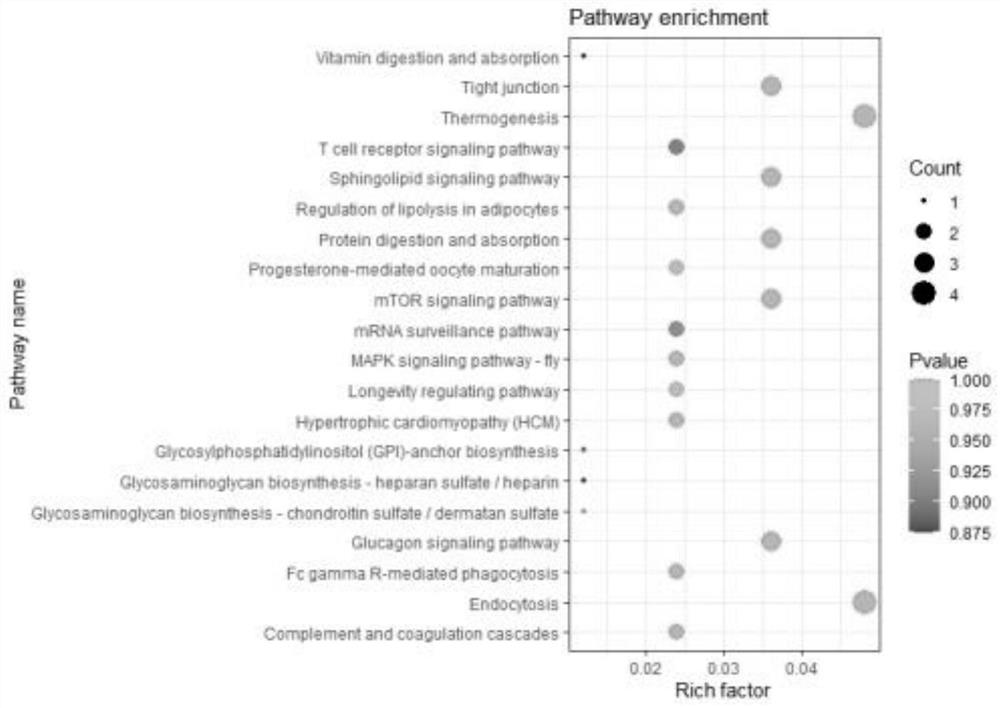
[0058] Example 2 Sequencing and Data Analysis
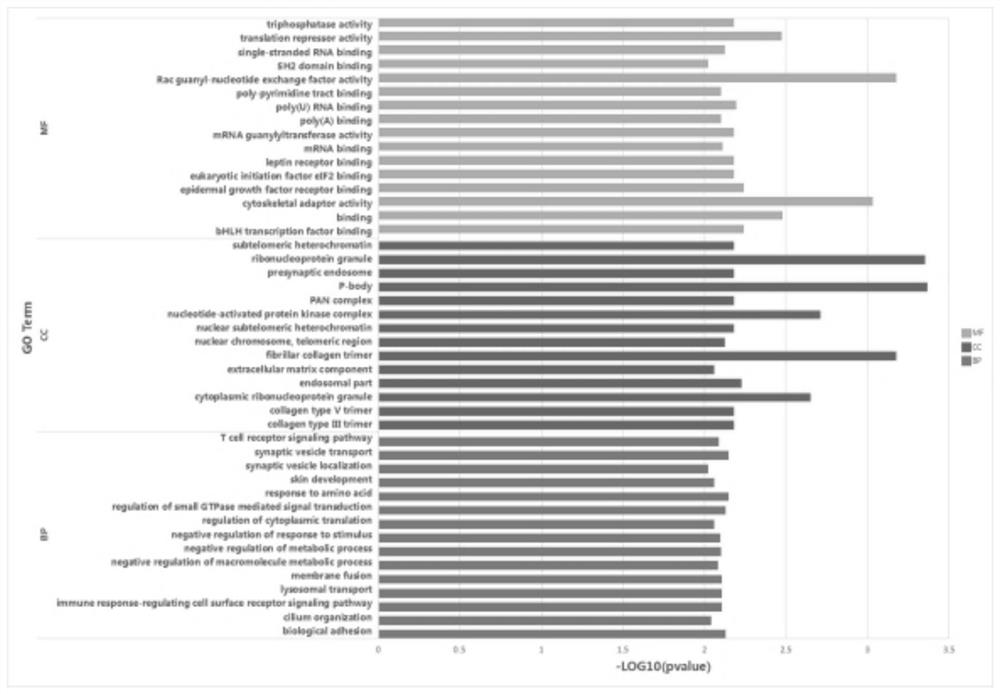
[0059] Sequenced using the Illumina NovaSeq 6000 sequencing platform. circRNA is a closed ring, which is formed by the reverse connection of the exon splicing acceptor and the donor site located upstream and downstream respectively. more stable. Linear RNA fragments can be aligned by linear alignment software, but Reads at the trans-splicing position of circular RNA cannot be directly aligned to the genome. Based on Back splicejunction (BSJ) reads, use related software (CIRI2+find_circ take union) to predict circRNA, and compare with database circBase (animal).
[0060] Classification is based on the start and end points of circRNAs on the genome. The first type of termination point and start point are located in the exon region of the gene as exon circRNA. The second type of starting point or ending point is located in the intron region of the gene as intron circRNA. When one end of the third type of starting point or endin...
Embodiment 3
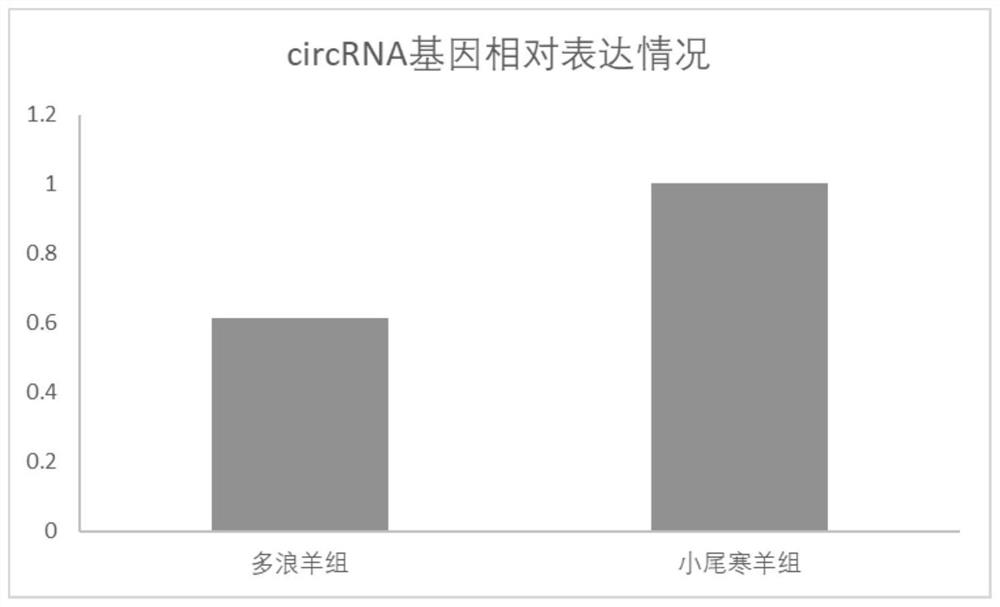
[0066] Embodiment 3 real-time fluorescent quantitative PCR detection verification
[0067] In this experiment, 20 Duolang sheep and Small-tailed Han sheep were collected, and the differentially expressed circRNA was detected by PCR between the two groups. Each gene was replicated three times to verify the differential expression of the NC_040256.1_12467238_12540333 gene.
[0068] The main method is as follows:
[0069] Use circRNA cDNA Synthesis Kit (GenePool, Cat#GPQ1805) for reverse transcription, the reaction system is: 2×FastSYBR Mixture, 10 μl; downstream primer (10uM) and upstream primer (10uM) each 0.4 μl; template, 2 μl; sterilized distilled water to 20 μl. See the table for primer information.
[0070] Table 1 PCR reaction results
[0071]
[0072] To export the data after the reaction, use 2 -ΔΔCt The relative expression of the samples was calculated by the method, and the t-test was used to analyze the relative expression. The data were shown as mean ± standa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com