Methods and compositions for the specific inhibition of gene expression by double-stranded RNA
a technology of double-stranded ribonucleic acid and effector molecules, which is applied in the field of compositions and methods for genespecific inhibition of gene expression by double-stranded ribonucleic acid (dsrna) effector molecules, can solve the problems of complex rules, unable to target sirna molecules, and early attempts to similarly suppress gene expression using long dsrnas in mammalians systems failed, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0050] This example demonstrates the preparation of double-stranded RNA oligonucleotides
[0051] Oligonucleotide synthesis and purification. RNA oligonucleotides were synthesized using solid phase phosphoramidite chemistry, deprotected and desalted on NAP-5 columns (Amersham Pharmacia Biotech, Piscataway, N.J.) using standard techniques (Damha and Olgivie, Methods Mol Biol 1993, 20:81-114; Wincott et al., Nucleic Acids Res 1995, 23:2677-84). The oligomers were purified using ion-exchange high performance liquid chromatography (IE-HPLC) on an Amersham Source 15Q column (1.0 cm×25 cm) (Amersham Pharmacia Biotech, Piscataway, N.J.) using a 15 min step-linear gradient. The gradient varied from 90:10 Buffers A:B to 52:48 Buffers A:B, where Buffer A was 100 mM Tris pH 8.5 and Buffer B was 100 mM Tris pH 8.5, 1 M NaCl. Samples were monitored at 260 nm and peaks corresponding to the full-length oligonucleotide species were collected, pooled, desalted on NAP-5 columns, and lyophilized.
[0052]...
example 2
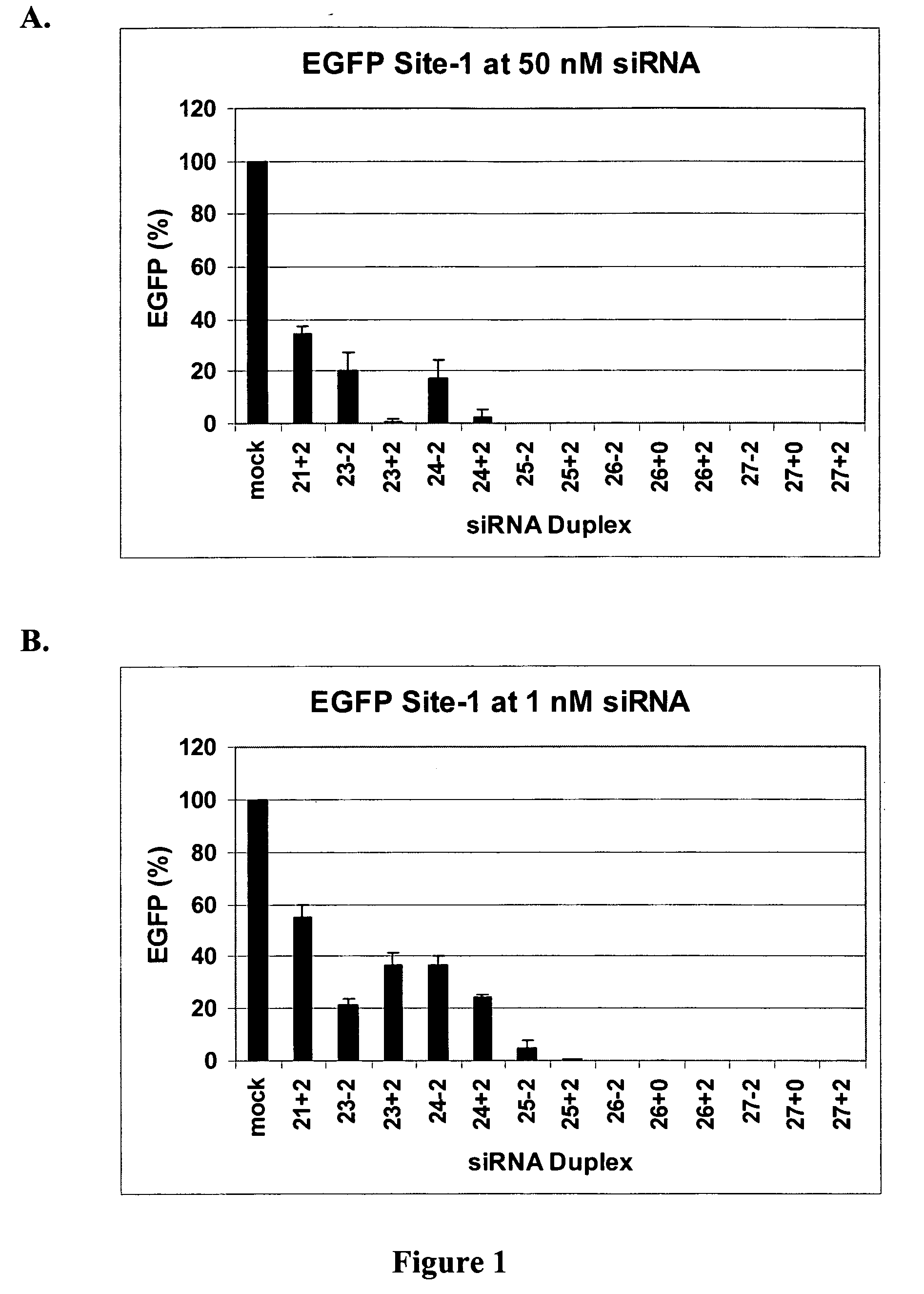
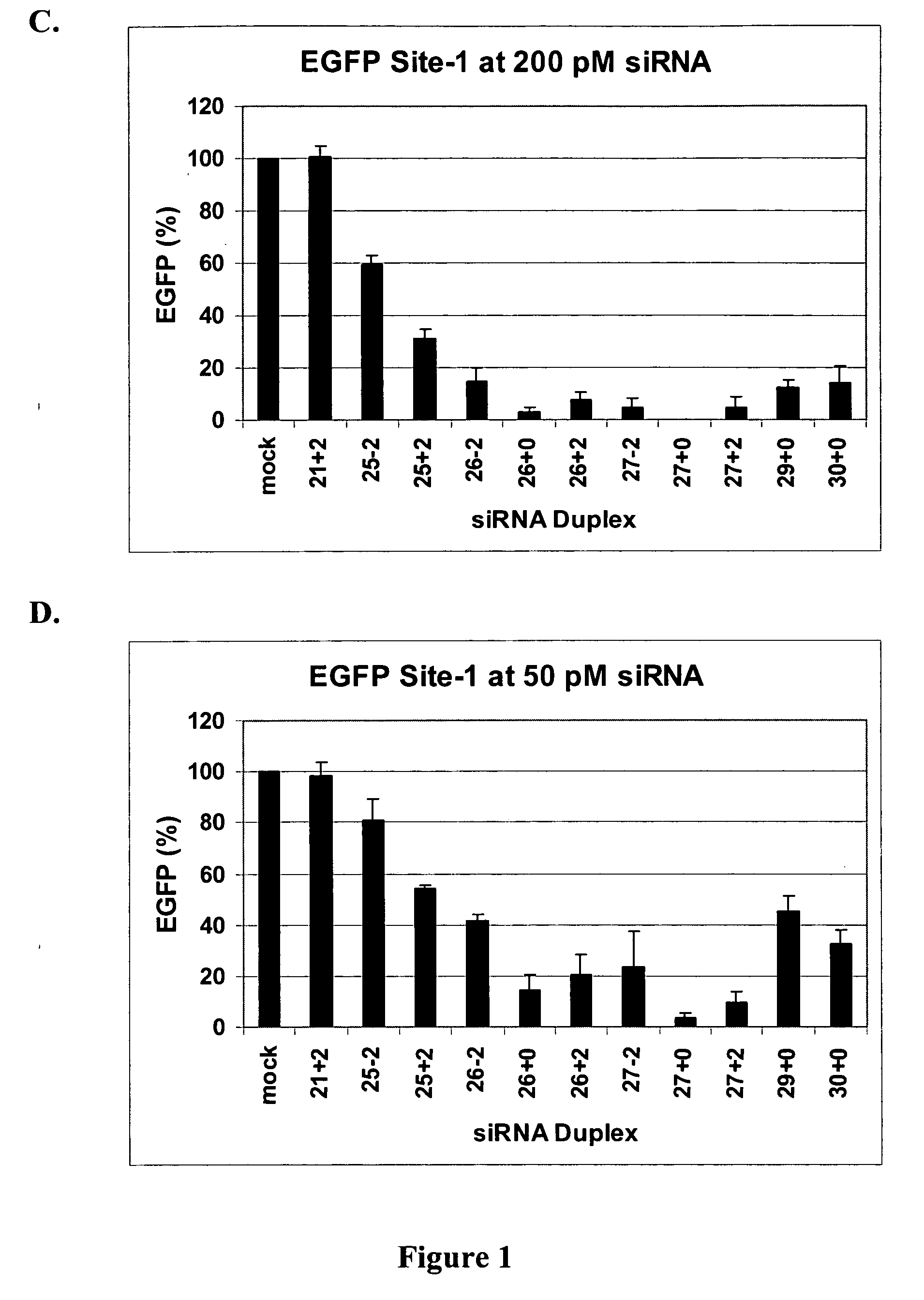
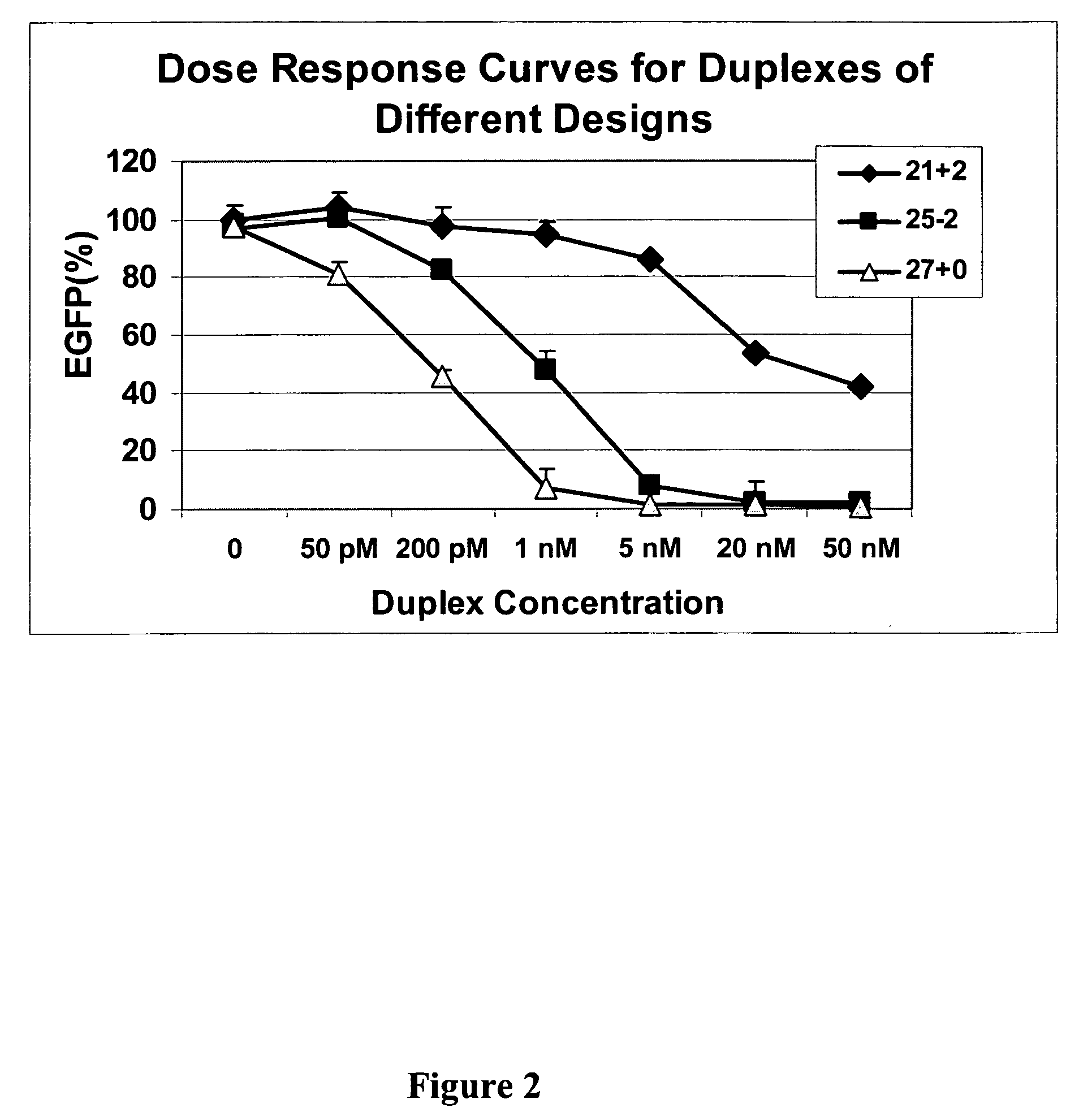
[0055] This example demonstrates that dsRNAs having strands that are 25 nucleotides in length or longer have surprisingly increased potency in mammalian systems than known 21-23-mer siRNAs.
[0056] Cell Culture, Transfection, and EGFP Assays. Human embryonic kidney (HEK) 293 cells were grown in DMEM medium supplemented with 10% fetal bovine serum (FBS) (Irvine Scientific, Santa Ana, Calif.). Transfections were done at 90% confluence in 24-well plates using Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Briefly, 50 μl of Opti-MEM media was mixed with nucleic acids, including siRNA duplexes and / or 100-200 ng plasmid pEGFP-C1 (Clontech, Palo Alto, Calif.) for 5 min. Nucleic acids were then mixed with 50 μl of Opti-MEM media that had been pre-mixed with 1.5 μl of Lipofectamine 2000 and incubated at room temperature for 15 min. The lipid—nucleic acid mixtures were added to cells after removal of old media and swirled and then an additional ...
example 3
[0068] This example demonstrates that the use of 25-30 nucleotide RNA duplexes allows gene targeting at a site that could not be effectively targeted using traditional siRNA 21 -mer designs.
[0069] It is currently expected in the art that the majority of 21-mer siRNA duplexes targeted to sites within a given target gene sequence will be ineffective (Holen et al., 2002, Nucleic Acids Res., 30:1757-1766). Consequently, a variety of sites are commonly tested in parallel or pools containing several distinct siRNA duplexes specific to the same target with the hope that one of the reagents will be effective (Ji et al., 2003, FEBS Lett., 552:247-252). To overcome the need to pool or engage in large scale empiric testing, complex design rules and algorithms have been devised to increase the likelihood of obtaining active RNAi effector molecules (Schwarz et al., 2003, Cell, 115:199-208; Khvorova et al., 2003, Cell, 115:209-216; Ui-Tei et al., 2004, Nucleic Acids Res., 32:936-948; Reynolds et...
PUM
| Property | Measurement | Unit |
|---|---|---|
| weight | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com