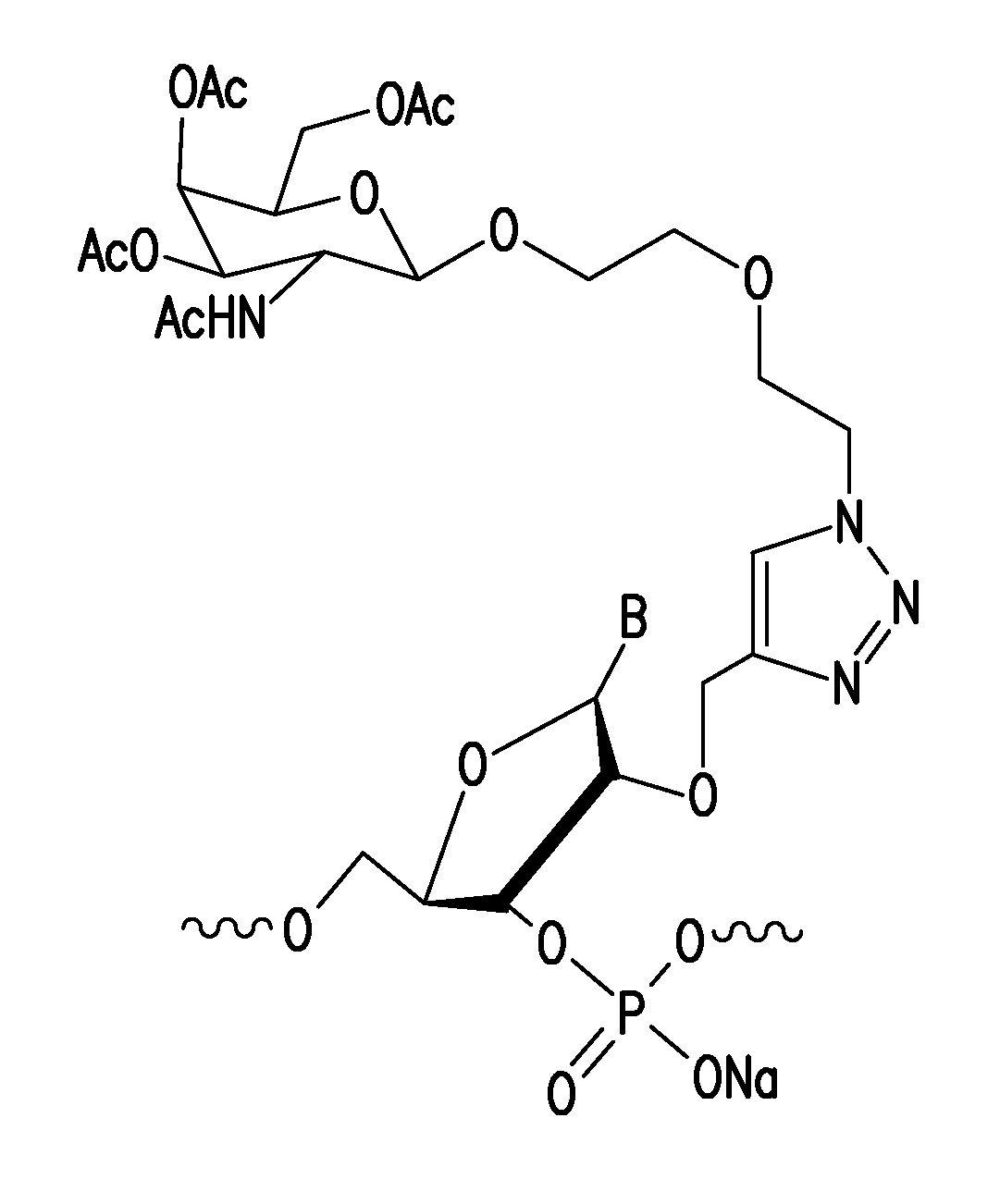
Post-synthetic chemical modification of RNA at the 2'-position of the ribose ring via "click" chemistry
a ribose ring and chemical modification technology, applied in the field of post-synthetic can solve the problems of low-throughput chemical modification of rna prior to synthesis, bear sensitive functional groups, and limited scop
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
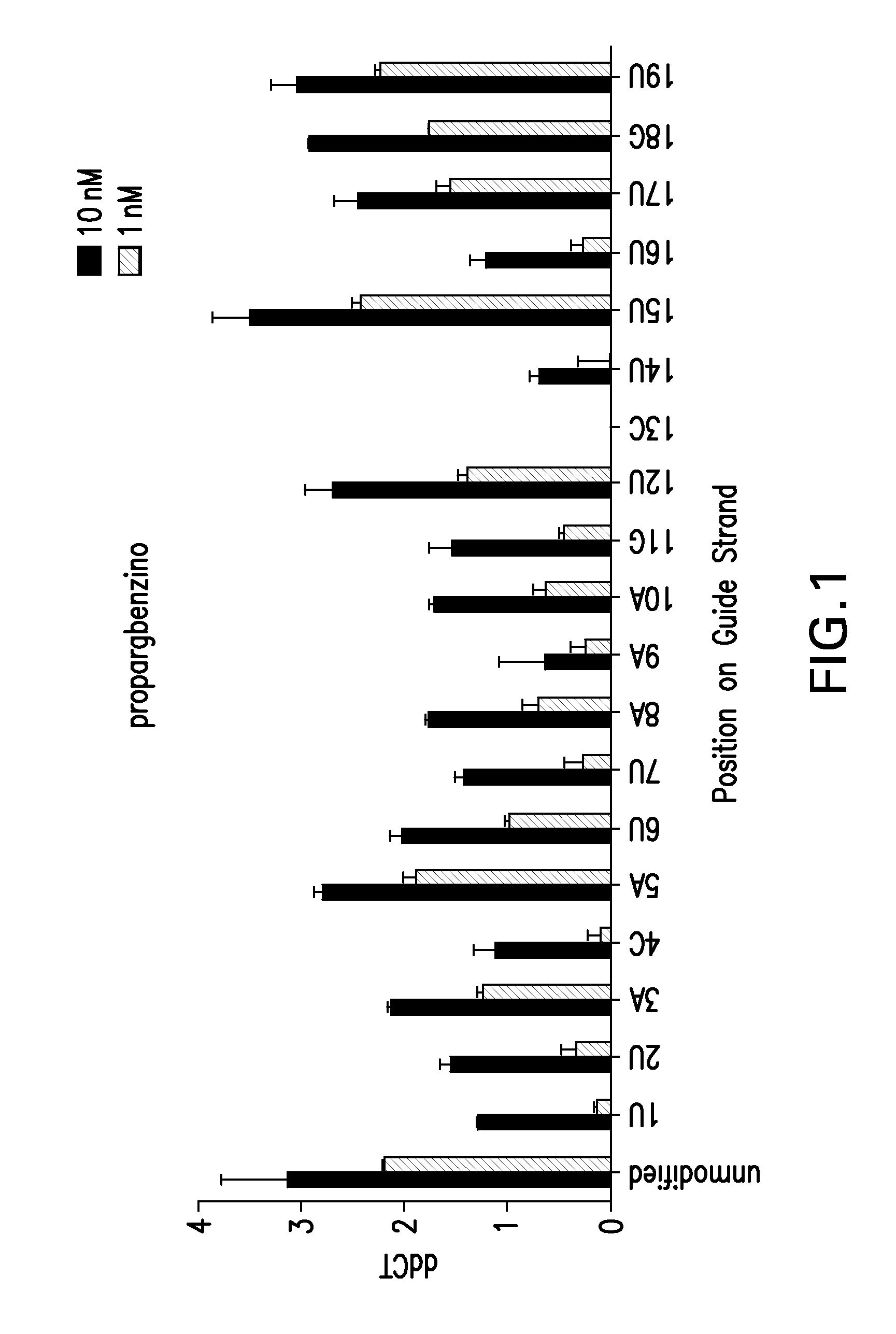
[0231]In FIG. 1, the impact on knockdown of the 2′-O-benzyl-triazole inosine chemical modification was systematically evaluated along positions 1 through 19 of the guide strand of an siRNA targeting mRNA SSB(291).
example 2
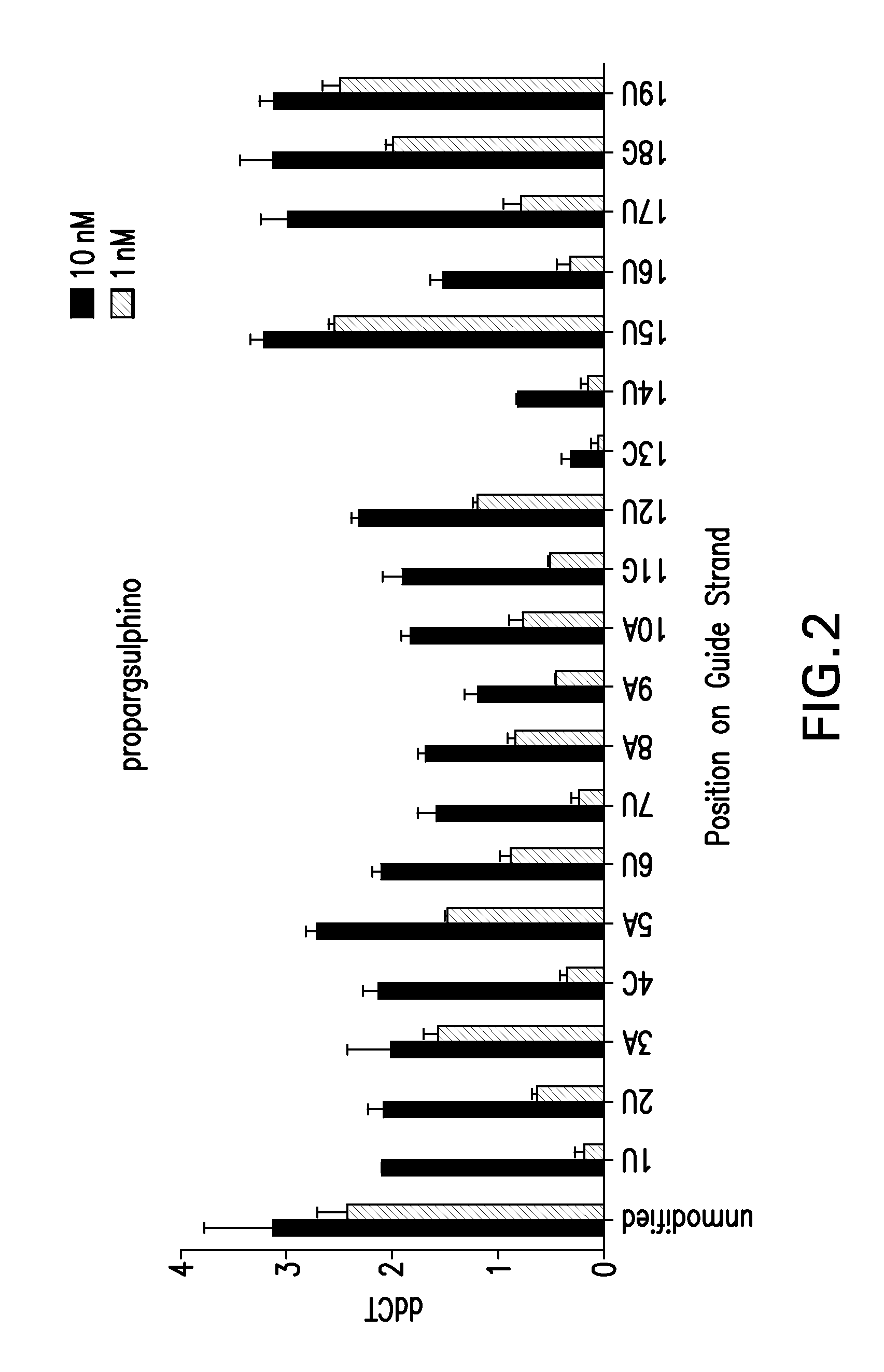
[0232]In FIG. 2, the impact of the 2′-O-phenylthiomethyl-triazole inosine chemical modification was systematically evaluated along positions 1 through 19 of the guide strand of an siRNA targeting mRNA SSB(291).
example 3
[0233]In FIG. 3, the impact on knockdown of the 2′-O-benzyl-triazole inosine chemical modification was systematically evaluated along positions 1 through 19 of the guide strand of an siRNA targeting mRNA Luc(80).
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com