Resistance gene Pi37 against rice blast and application thereof
A rice blast resistance gene technology, applied in the field of cloning of rice blast resistance gene Pi37, can solve the problems of cloning rice blast resistance genes, etc., and achieve the effect of shortening the breeding time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
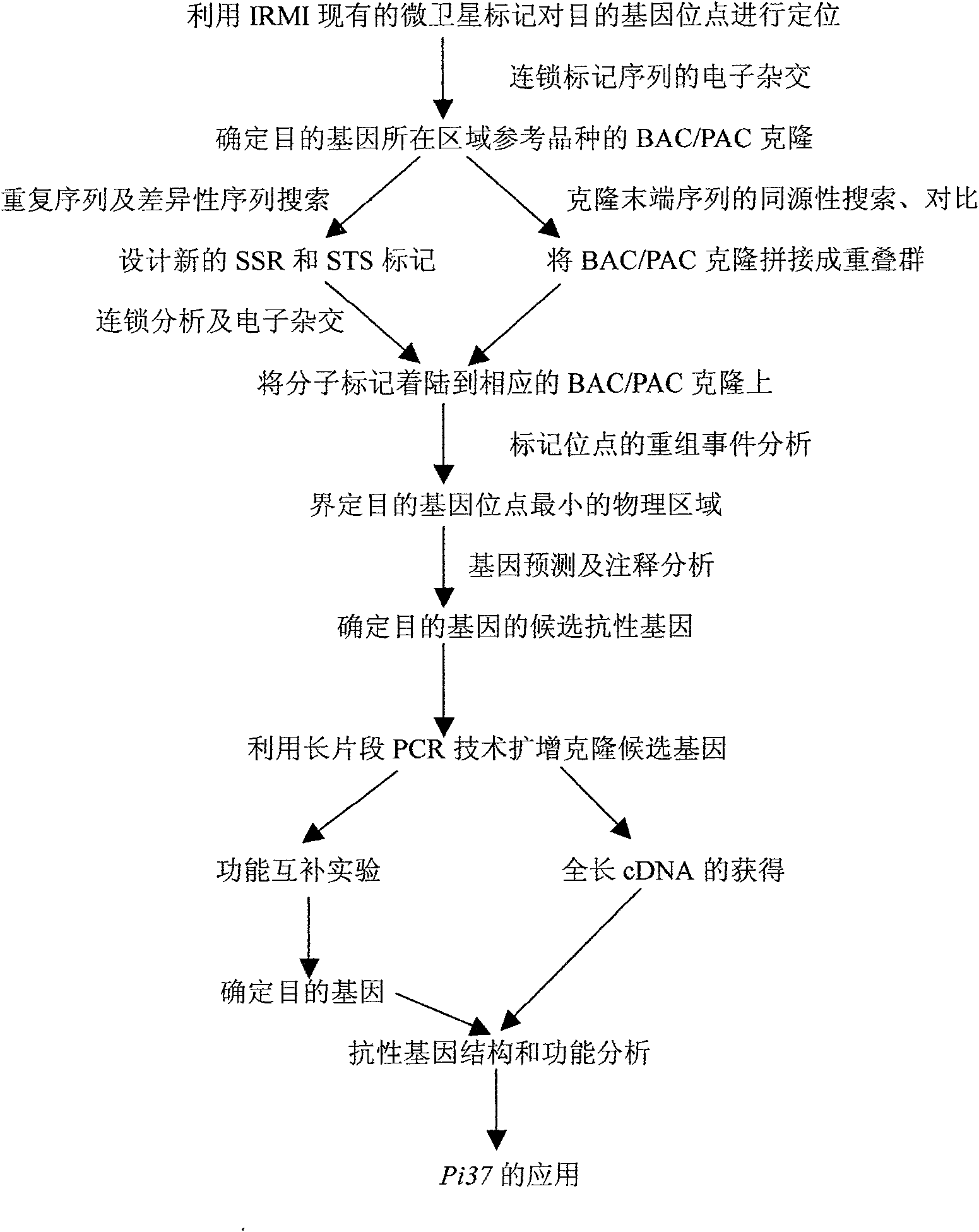
[0039] Example 1: Genetic Analysis and Preliminary Mapping of Rice Blast Resistance Gene Pi37
[0040] The present invention is for 3 F which are derived from the hybrid combination of japonica rice disease-resistant variety Q1333 and 3 indica rice disease-susceptible varieties AS20-1, C101PKT and CO39. 2 Populations were inoculated with strains CHL1159, CHL1340 and CHL1405 that showed distinct non-affinity / affinity responses to the parental varieties, and the results showed that the three F 2 The segregation ratios of resistant plants and susceptible plants in the population were all consistent with 3R:1S, so it was inferred that the resistance of Q1333 to the above three inoculated strains was controlled by a pair of dominant genes. to these 3 F's 2 The linkage analysis of the resistance genes corresponding to the population showed that they were controlled by the same dominant gene, so the three F 2 From the population, a large mapping population consisting of 85 resistan...
Embodiment 2
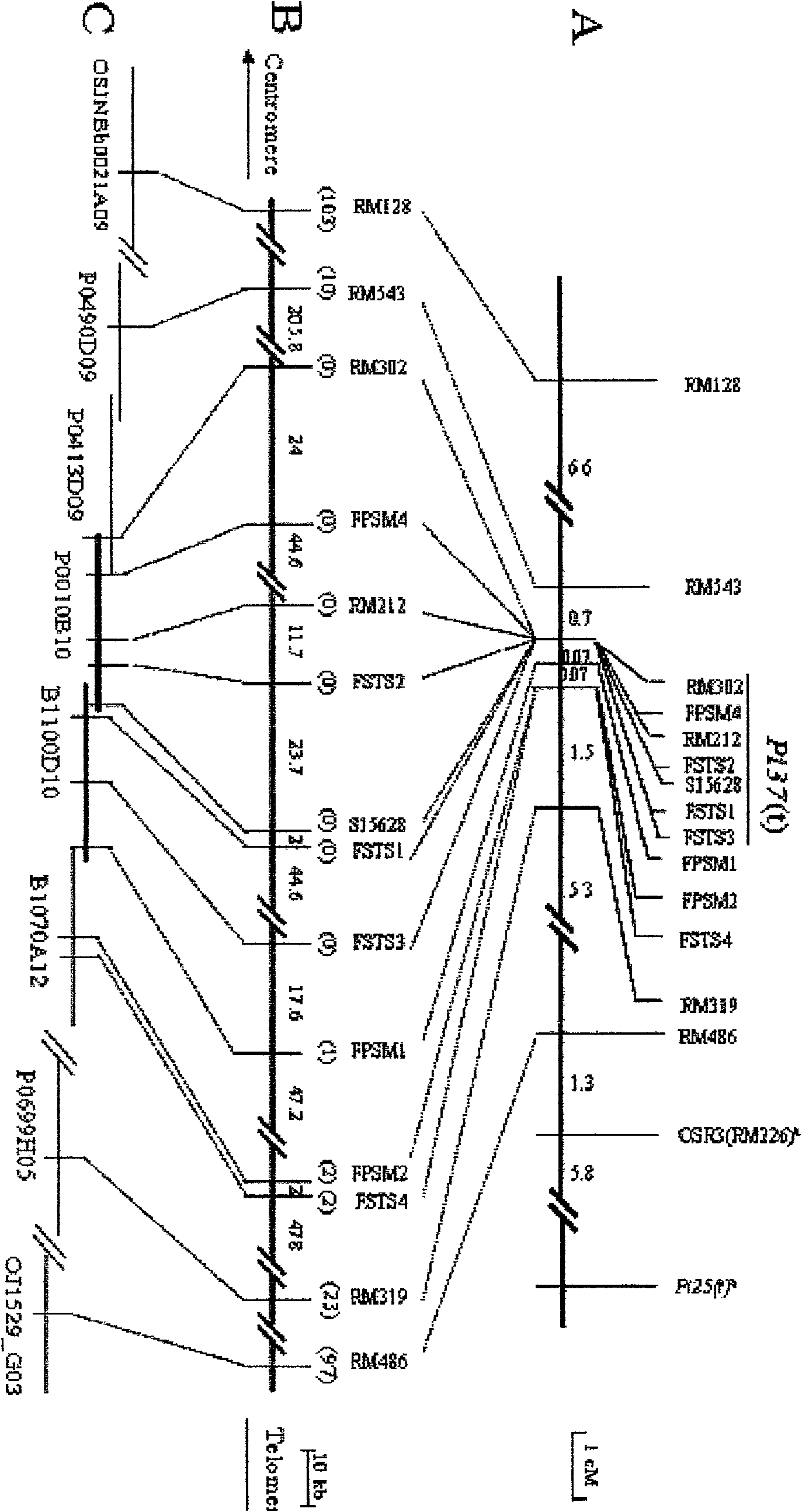
[0042] Example 2: Fine Mapping of Rice Blast Resistance Gene Pi37
[0043] In order to narrow down the Pi37 locus to a smaller genomic region, the reference sequence of Nipponbare (Nipponbare) was sequenced using the public database, and new molecular markers were developed in the target genomic region by bioinformatics analysis (BIA). Namely the new SSR and STS tags. Among the 8 markers developed FPSM1, FPSM2, FPSM3, FPSM4, FSTS1, FSTS2, FSTS3 and FSTS4, only marker FPSM3 showed no polymorphism; further analysis of recombination events showed that FPSM1 detected 1 recombinant, FPSM2 and FSTS2 Two recombinants were detected, and the other four markers FPSM4, FSTS1, FSTS3 and FSTS4 were completely co-segregated with the target gene. Based on the results of the above linkage analysis, the Pi37(t) locus is defined in the genetic region of 0.77cM between the flanking markers RM543 and FPSM1, and there are seven molecular markers RM302, FPSM4, RM212, FSTS4, S15628, and FSTS1 in th...
Embodiment 3
[0045] Example 3: Transformation of rice blast new resistance gene Pi37 and resistance identification of transformants
[0046] In order to determine the functions of the candidate resistance genes, functional complementation experiments were carried out on the four predicted candidate genes R37-L1, R37-L2, R37-L3 and R37-L4. Four candidate resistance genes were amplified by long-range PCR (LR-PCR) technology and cloned into the binary vector transformation system pCAMBIA1300, which was introduced into Agrobacterium strain EHA105. The mature seeds of the highly susceptible variety Q1063 were used to induce callus on the induction medium. The EHA105 containing the target gene transformation vector was cultured in YM agar medium at 28°C for 1 day, and collected in MB liquid medium containing 100 μmol / L acetosyringone. The rice callus was immersed in the bacterial solution for 20 minutes, blotted dry and transferred to MB agar medium for culture. Transfer the callus to the cult...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com