Methods for production of oligonucleotides
一种寡核苷酸、核苷酸的技术,应用在用于产生寡核苷酸领域,能够解决不提供待扩增DNA序列末端自由设计等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0142] Preparation of probe (A) (step a)
[0143] The starting nucleic acid sequence (probe (A)) comprising one or more oligonucleotides to be amplified and one or more nicking cassettes can be synthesized by standard chemical methods, such as β-cyanoethylphosphoramidite Chemical. A 5'-phosphate can be added during this synthesis, or alternatively a 5'-phosphate can be enzymatically coupled to the 5'-terminus of the nucleic acid sequence, for example by using T4 polynucleotide kinase.
[0144] Long sequences can be constructed by external templated ligation of multiple oligonucleotides, eg when the oligonucleotides to be amplified are very long, eg 200-1000 nucleotides. In this case, probe (A) can be prepared by external templated ligation of two or more oligonucleotides (Fig. 5). In one embodiment, the invention relates to a method wherein probe (A) is prepared by ligating one or more nucleic acid sequences comprising one or more of said one or more oligonucleotides A plur...
Embodiment 1
[0421] Amplification of the 66 nucleotide oligonucleotide SF86-SC by using the suicide cassette
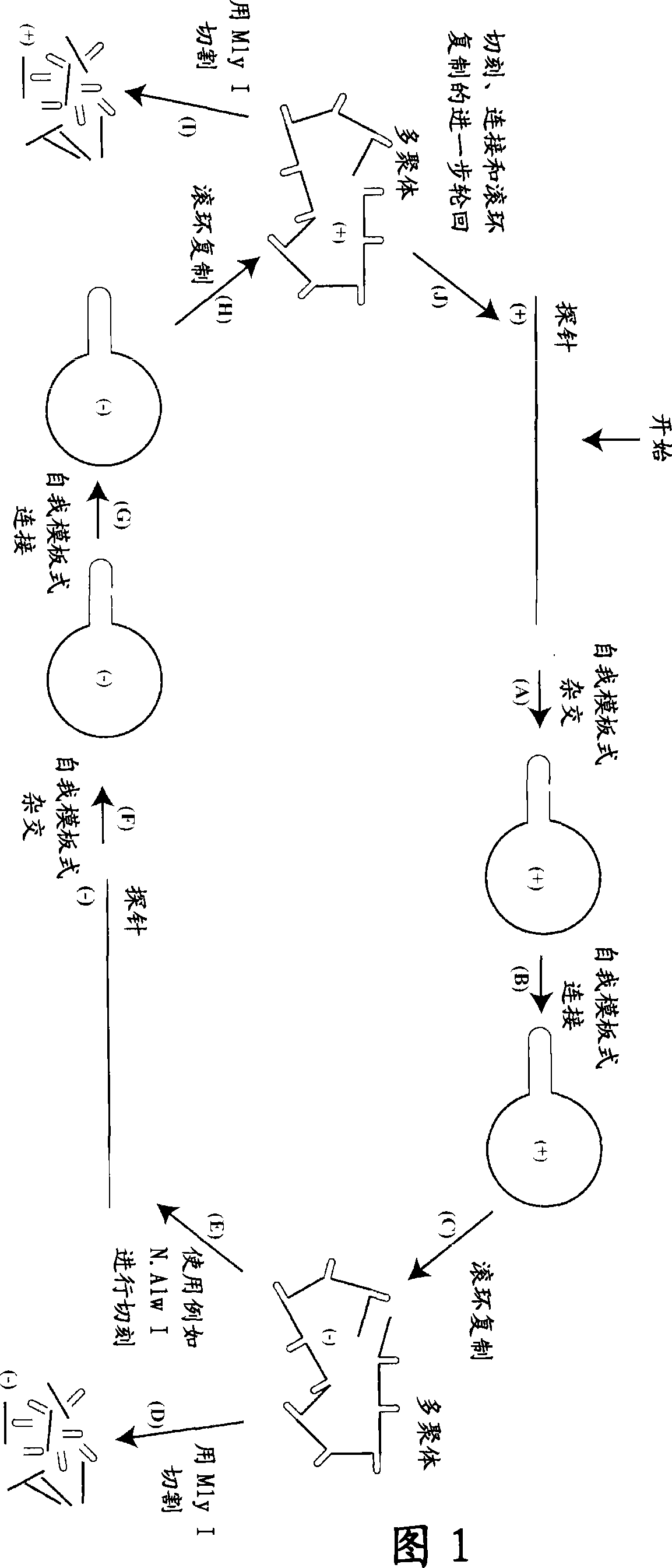
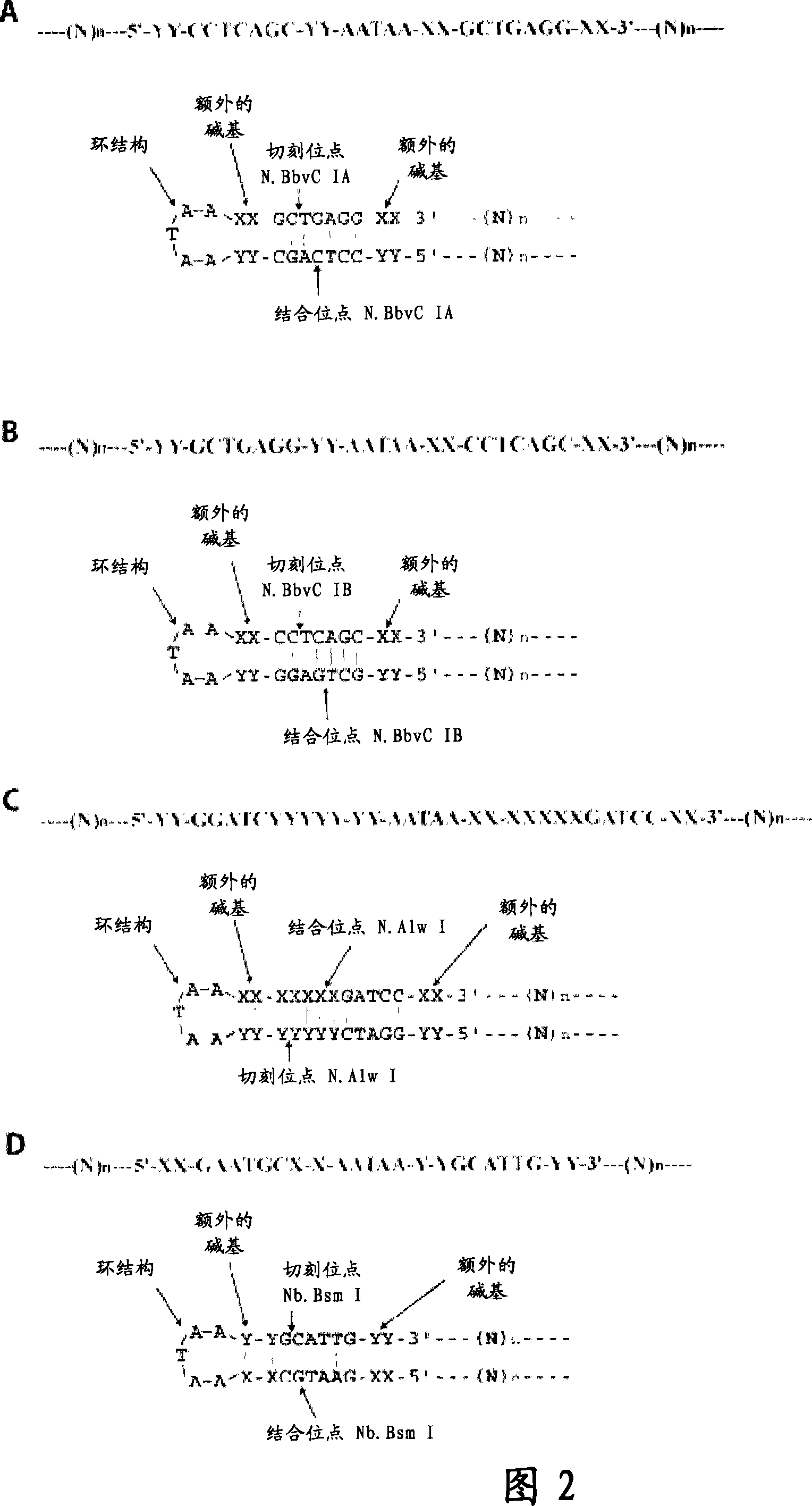
[0422] By using the present invention, we were able to amplify oligonucleotides approximately 100,000-fold. The present invention uses a suicide cassette as a tool to amplify specific oligonucleotides. The suicide cassette is a hairpin structure containing binding sites for the nicking and restriction enzymes; in this example, the nicking and restriction enzymes are N.Alw I (Nt.Alw I) and Mly I, respectively (Fig. 3, A-B). A linear DNA sequence can be turned into a closed circular sequence by self-templated ligation by placing a suicide cassette at each end of the initially synthesized oligonucleotide. Oligonucleotides can be amplified at either polarity by successive rounds of rolling circle replication, nicking and ligation. In a final step, the suicide cassette can be separated from the oligonucleotide by restriction cleavage with Mly I (Figure 1).
[0423] Oligonucleotide ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com