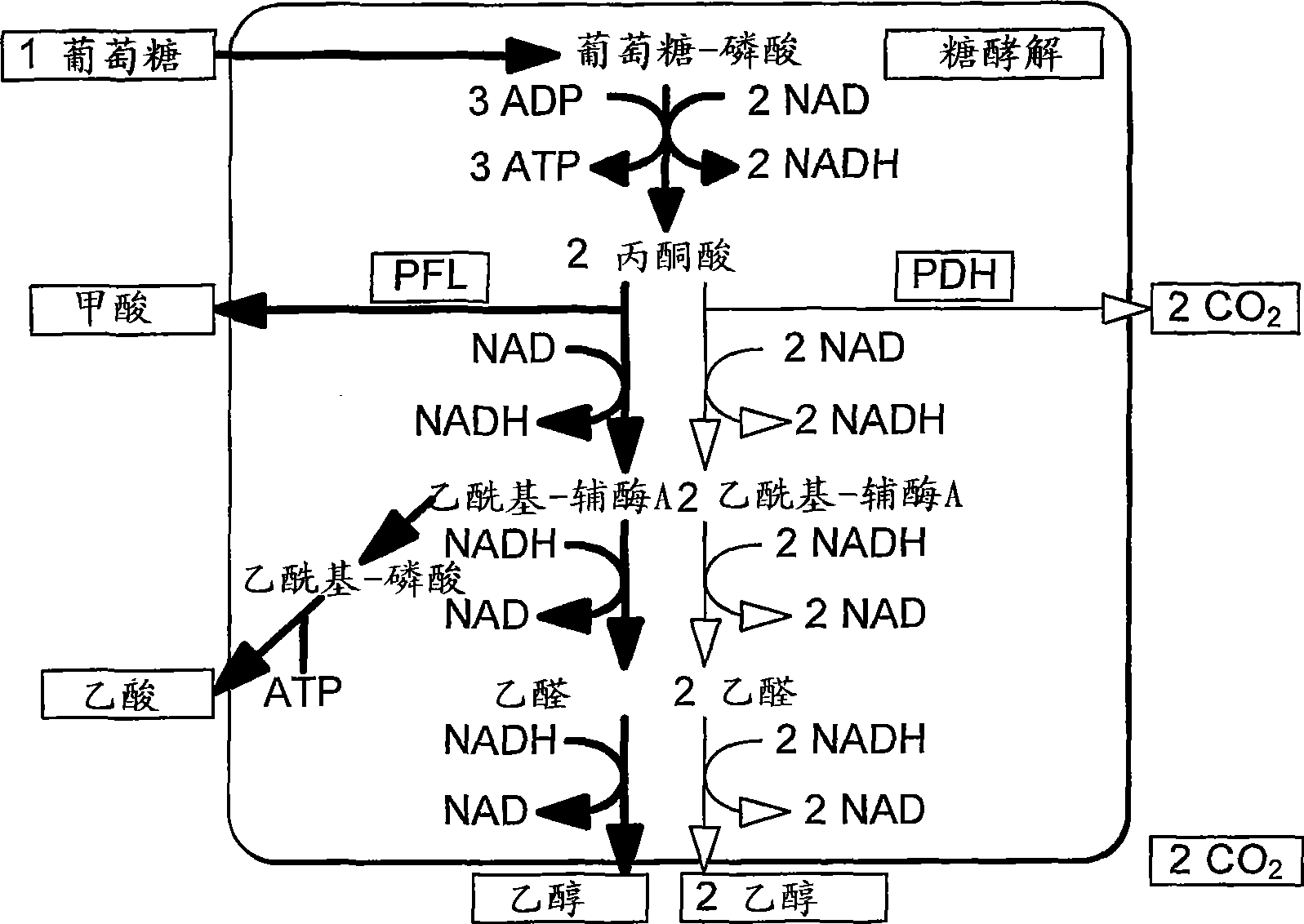
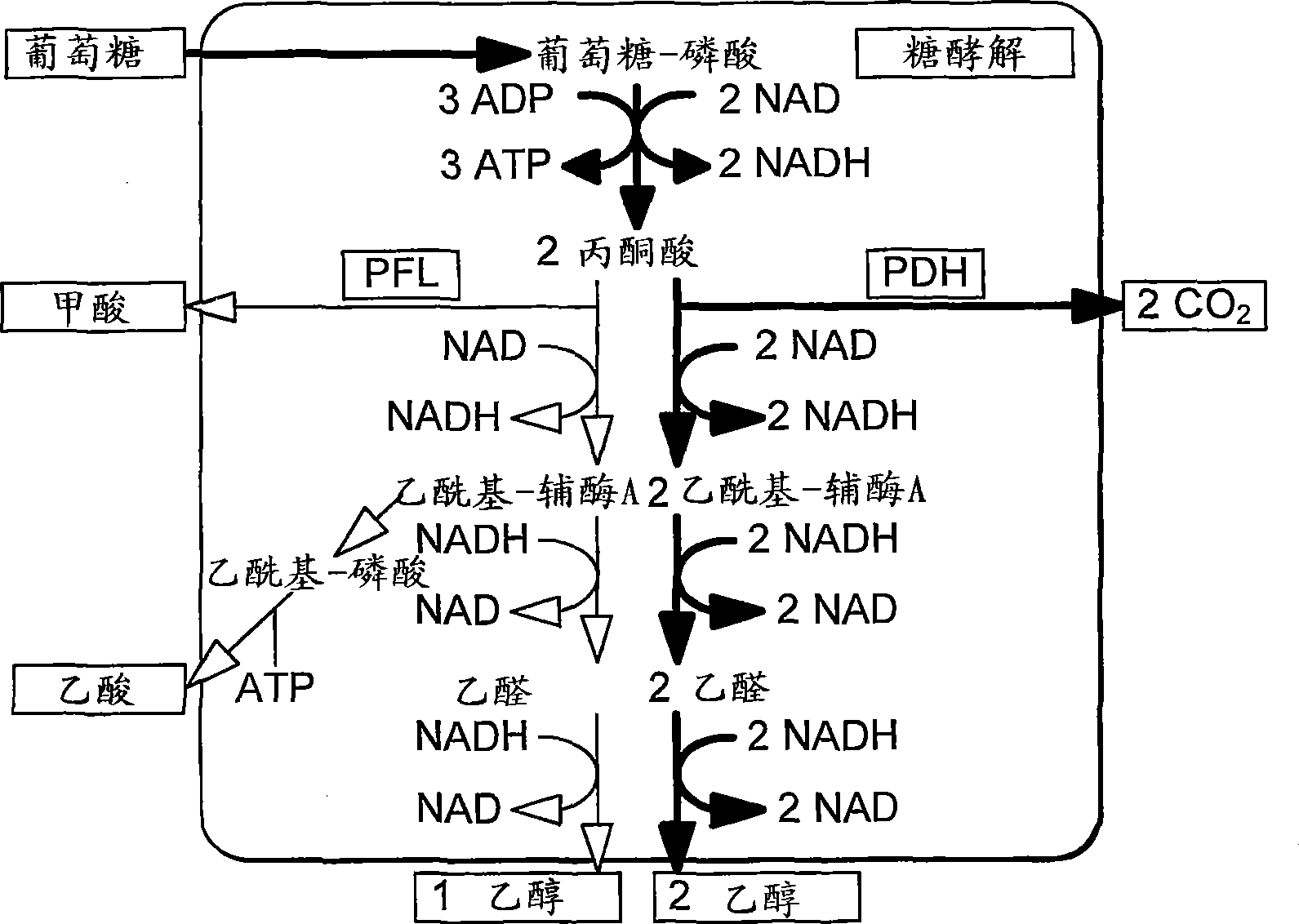
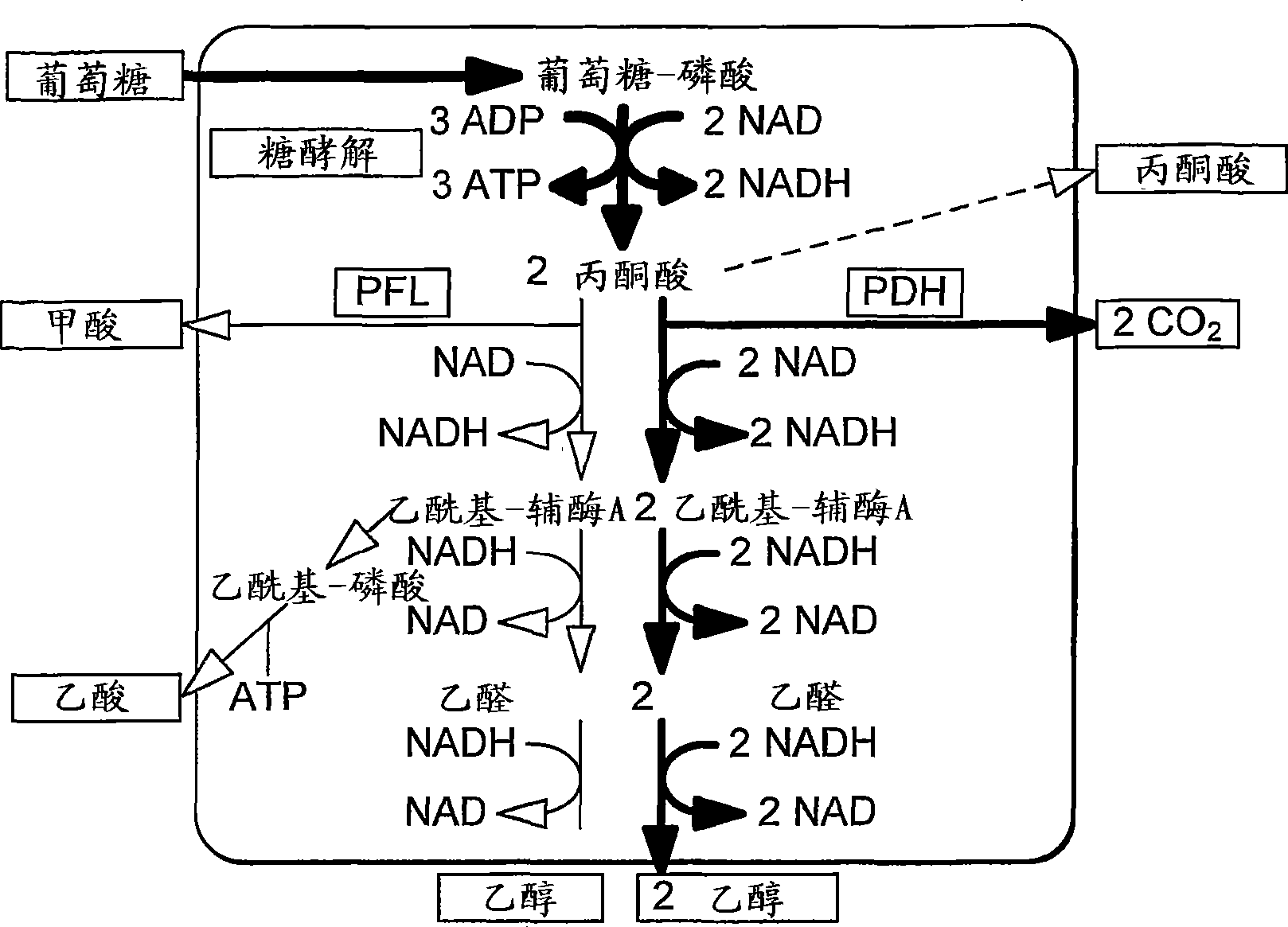
Enhancement of microbial ethanol production
A technology of microorganisms, thermophilic microorganisms, applied in the field of microorganisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Embodiment 1. Construction of synthetic formate dehydrogenase gene ( figure 2 )
[0077] Amino acid sequence of Pseudomonas 101 formate dehydrogenase (NCBI protein database accession number P33160 - SEQ ID NO: 3) back translated into DNA sequence by codon optimization for Geobacillus thermoglucosidase . The promoter and rho-independent terminator regions from the Bacillus strain were added upstream and downstream of the translated sequence ( figure 2 ). The new sequence has less than 40% similarity to the known fdh gene sequence (37% identity to the known fdhl gene). Xba1 sites were designed on both sides of this construct to facilitate cloning into a suitable vector.
[0078] Using the method of Gao et al. (see Xinxin Gao, Peggy Yo, Andrew Keith, Timothy J.Ragan and Thomas K.Harris (2003). Nucleic Acids Research, 31(22), e143) to synthesize the required sequence, and clone it into pCR - Unique Xba1 location in Blunt. The resulting vector pCR-F1 ( Figure 4 )...
Embodiment 2
[0080] Example 2. Insertion of the fdh gene into multiple (IS) sites
[0081] This strategy works for some strains, such as the Bacillus stearothermophilus LLD-R strain, which contain insertion sequences (IS) that frequently recombine at multiple insertion sites. It is expected that a vector carrying the fdh gene and this IS sequence will be stably integrated at one or more of these positions.
[0082] Construction of plasmid pUB-ISF1 ( Figure 5 )
[0083] First, using Bacillus stearothermophilus LLD-15 as a template, use forward primer (AGTACTGAAATCCGGATTTGATGGCG-SEQ ID NO: 6) and reverse primer (AGTACTGCTAAATTTCCAAGTAGC-SEQ ID NO: 7) to PCR amplify the known LLD-R strain Insertion sequence (SEQ ID NO: 5 and image 3 ). Sca1 restriction sites were introduced at both ends of this sequence. The PCR product was first cloned into plasmid pCR-TOPO2.1 and the resulting plasmid pCR-IS was introduced into E. coli DH5α cells and used to isolate the IS region by Sca1 restrict...
Embodiment 3
[0088] Example 3. Construction of ldh-deleted strains.
[0089] The first step involves cloning the Bacillus kanamycin resistance marker (kan) and the gene cassette for the ldh gene of the Bacillus stearothermophilus LLD-R strain into plasmid pUC18, which replicates only in Gram-negative microorganisms .
[0090] Construction of Bacillus cloning vector plasmid pUCK ( Image 6 ).
[0091] The kanamycin resistance gene (kan) was cloned into the unique Zra1 site in plasmid pUC18, which is outside of any coding region and reporter gene (lacZ) in the plasmid. To clone the kan gene, use the plasmid pUB110 as a template, and use the following primers to amplify a 1.13kb fragment containing the kanamycin resistance gene by PCR:
[0092] kan-Bs Z-F (ACACAGACGTCGGCGATTTGATTCATAC-SEQ ID NO: 10) and
[0093] kan-Bs Z-R (CGCCATGACGTCCATGATAATTACTAATACTAGG-SEQ ID NO: 11).
[0094] The Zra1 sites were introduced into both ends of the kan gene by primers. The PCR product was then di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com