DNA sequence similarity detecting method based on Hurst indexes
A DNA sequence and detection method technology, applied in the direction of electrical digital data processing, special data processing applications, instruments, etc., to achieve the effect of simplifying the calculation complexity, concise method, and improving the degree of discrimination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
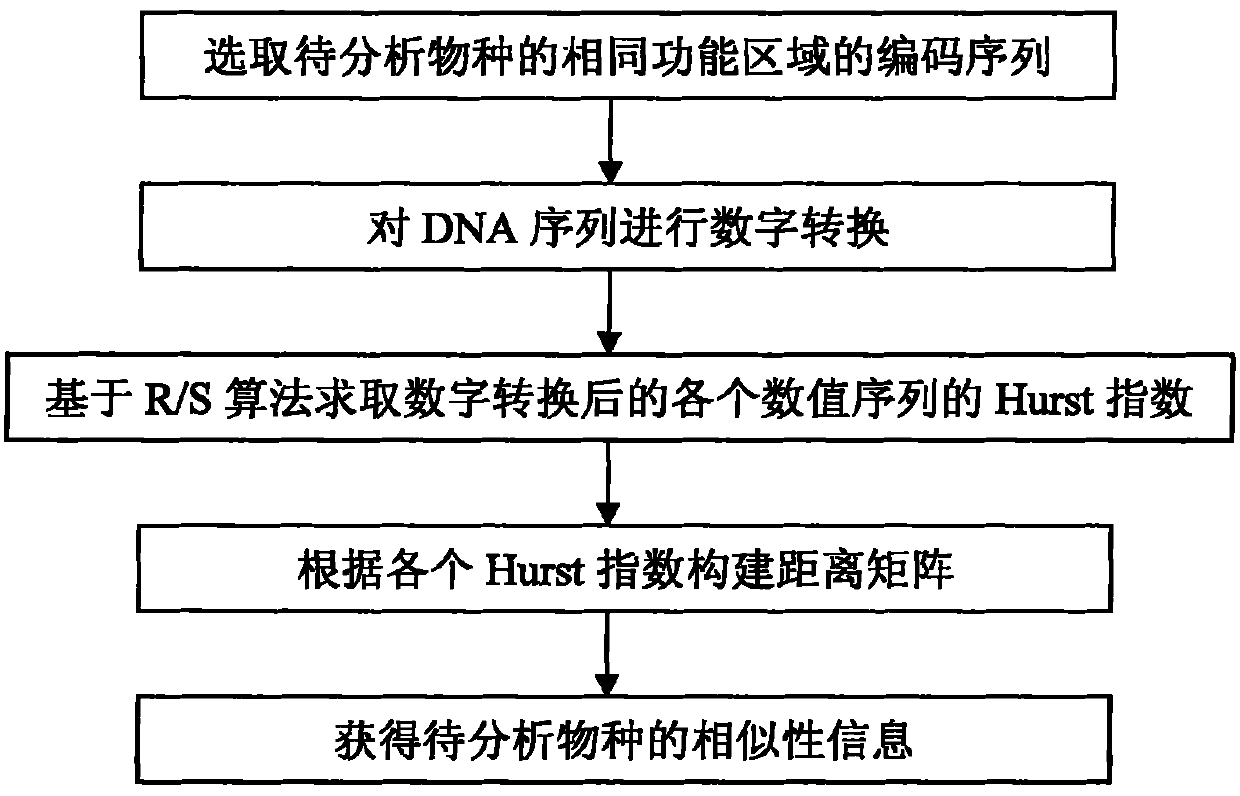
[0028] With reference to the accompanying drawings, the first exon coding sequence of the beta-globulin used in 11 species by the method of the present invention will be described in detail as the analysis object as an example, including the following steps: see figure 1 , the DNA sequence similarity detection method based on the Hurst index of the present embodiment comprises the following steps:
[0029] 1) The coding sequence of the first exon of β-globin, which is widely used in the analysis of sequence similarity, was selected as the initial sequence; see Table 1.
[0030] 2) Digitally convert the initial sequence obtained in step 1) to obtain the corresponding numerical sequence of the initial sequence; this method adopts 2D graphic representation to digitize the DNA sequence (see M, M, N, et al. Chemical Physics Letters [J]., 2003, 368: 1-6.). The characteristic of this method is that the expression of 4 bases depends on the sequence in which they appear in the an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com