Establishing method of bifidobacterium functional gene no-trace knockout method
A bifidobacteria, functional gene technology, applied in microorganism-based methods, biochemical equipment and methods, bacteria, etc., can solve the problems of limiting research progress in the field of bifidobacteria genetic engineering, difficult gene manipulation, and cell wall thickness, etc. The effect of avoiding the application of antibiotics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
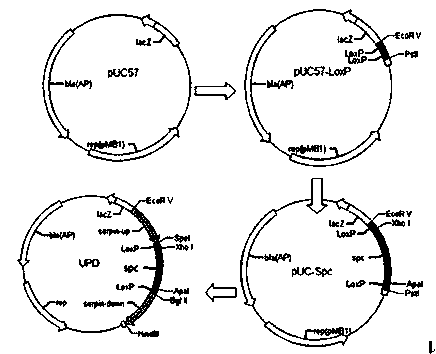
[0038] Example 1: Construction of Bifidobacterium Targeting Vector and Shuttle Expression Vector:
[0039] 1. In view of the characteristics of the LoxP site sequence, two LoxP sites with the same orientation were directly synthesized on the pUC57 vector, and the sequence (108 bp in total) was from Eco R V sites and Pst I site insertion, after which the spectinomycin resistance gene will be inserted from the middle and both ends of the two LoxP sites respectively spc and upstream and downstream homology arm sequences.
[0040] This step constructs the vector pUC57-LoxP. The synthetic sequences of the two LoxP sites are:
[0041] 5'-CGG ACTAGT ATAACTTCGTATAATGTATGCTATACGAAGTTAT CTCGAG AACGATCGGTT GGGCCC ATAACTTCGTATAATGTATGCTATACGAAGTTAT AGATCT CC-3'(SEQ ID.NO.1), the endonucleases are Speech I, xho I, Apa I, Bgl II.
[0042] 2. Using the pER8 plasmid as a template, design the primer P-Spc-F: 5’-CCG CTCGAG TGGTCCAGAACCTTGACCG-3'( xho I), (SEQ. ID....
Embodiment 2
[0051] Embodiment 2: the preparation and transformation of Bifidobacterium longum protoplast:
[0052] 1. Transformation of targeting vector: transfer Bifidobacterium longum NCC2705 from a glycerol tube stored at -70°C to an MRS plate (MRS agar medium: 10 g peptone, 10 g beef extract, 5 g yeast extract, Tween-80 1 g, K 2HPO 4 2 g CH 3 COONa·3H 2 O 5 g, triammonium citrate 2 g, MgSO 4· 7H 2 O 0.2 g, MnSO 4· h 2 O 0.05 g, Agar 15 g, glucose 20 g, add distilled water to 1000 mL, adjust the pH to 6.2-6.4, and autoclave at 121 °C for 15 min. MRS broth medium: the composition is the same as above without adding agar), cultured anaerobically at 37°C for 24 h. Pick a single colony from the plate, inoculate it in 5 mL MRS liquid medium, and culture it anaerobically at 37 °C for 24 h; then transfer it to fresh 25 mL MRS liquid medium , anaerobic culture at 37°C for 24 hours; centrifuge to collect the bacteria, resuspend; add mutanolysin to a final concentration of 4.5-5.5 mg / L...
Embodiment 3
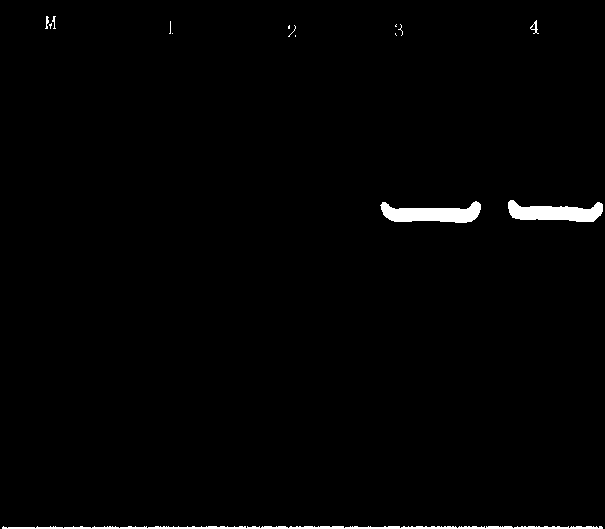
[0056] Example 3: Bifidobacteria serpin Verification of gene deletion strains:
[0057] 1. to ⊿serpin⊿spc strain genomic DNA as a template, amplified with primers serpin-F / R (SEQ. ID.NO.11 and SEQ. ID.NO.12) serpin Gene (amplification system and reaction procedure are the same as step 2 in Example 2), detected during recombination serpin Changes in gene signal; amplified with primers SEQ.ID.NO.2 and SEQ.ID.NO.3 spc Gene, detection of shuttle expression vector before and after transformation spc gene signal. Figure 10 It is the result of PCR verification after step-by-step transformation, after serpin PCR amplification of gene deletion fragments and 1% agarose gel electrophoresis, showing ⊿serpin The strain can amplify 1000 bp, indicating that the transformant genome contains spc Gene; ⊿serpin The strain failed to amplify the 1400 bp fragment, indicating that homologous recombination occurred between the targeting fragment and the homologous fragment on the c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com