Traceless fixed-point transformation method of large gene cluster and application of traceless fixed-point transformation method
A technology of fixed-point transformation and gene clustering, applied in the field of molecular biology, can solve the problems of low transformation efficiency of Saccharopolyspora spinosa and obstacles to the application of combinatorial biosynthesis technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
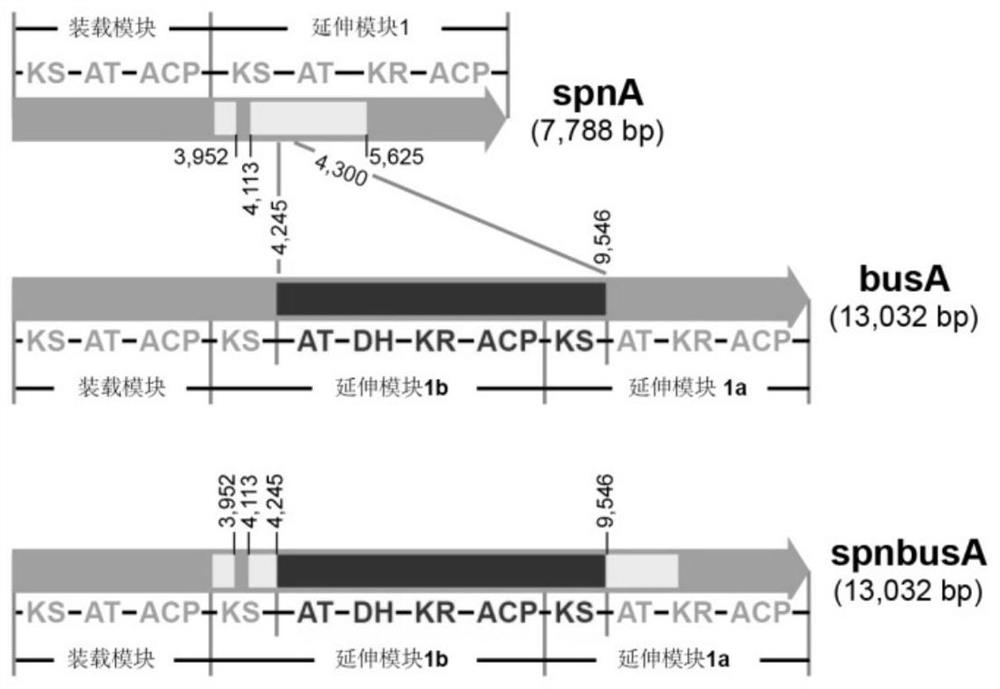
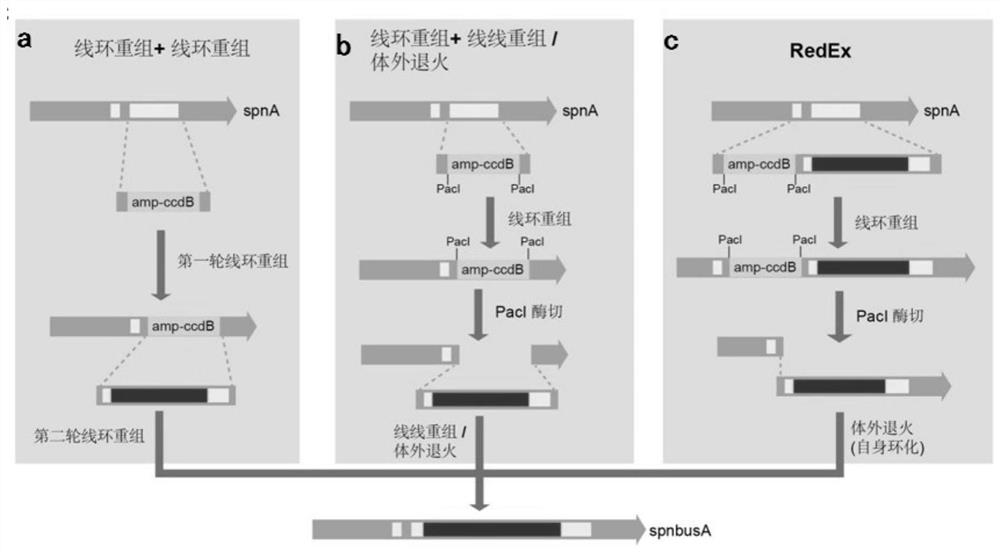
[0074] Example 1. Using RedEx technology to seamlessly insert the busA AT1b-KS1a domain into the spnA gene
[0075] 1. Preparation of NDA fragment ampccdB-busA20 carrying a 20bp terminal homology arm:
[0076] Use the pR6K-oriT-phiC31 plasmid as a template for PCR amplification. The primers for PCR amplification are as follows, and the underline is the BstZ17I restriction site:
[0077] R6K-2: 5'-AACGCGCTGCGTGAATCTTCCGCCGGCGACATGGGCAGGCGTGTCGAAGCGAAGTTCTGGGGCGCCGTCGAGCACGAAGA GTATAC AGTTCAACCTGTTGATAGTACG-3',
[0078] R6K-3: 5'-CCAGAAGTCGGCTCATCCACGTGCAACGTGCGCGGTAGCTGCCCGTGCCGCATCGCCATCACCATCTTCATGACGCCGGC GTATAC TGTCAGCCGTTAAGTGTTCCTGTG-3',
[0079] Then use the above PCR product as a template to carry out PCR amplification to obtain the R6K replicon. The primers for PCR amplification are as follows:
[0080] R6K-1: 5'-GCTGCCCACCTACGCCTTCCAACGACAGCGGTACTGGCTGAACGCGCTGCGTGAATCTTC-3',
[0081] R6K-3: 5'-CCAGAAGTCGGCTCATCCACGTGCAACGTGCGCGGTAGCTGCCCGTGCCGCATCGCCATCACCATCT...
Embodiment 2
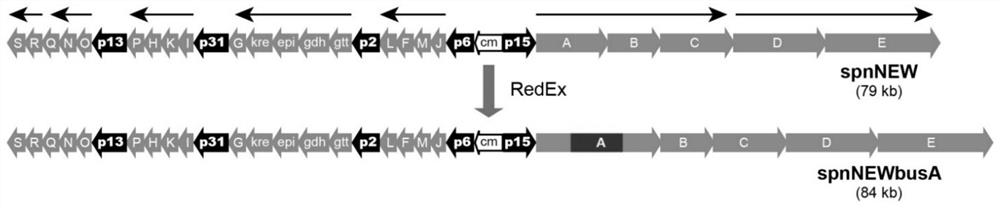
[0090] Embodiment 2, heterologous expression of butenyl spinosyn
[0091] 1. Product Analysis of Butenyl Spinosad by High Performance Liquid Chromatography-Mass Spectrometry (HPLC-MS)
[0092] The recombinant pBAC-spnNEWbusA was integrated into the attB site of Streptomyces albicans J1074 genome by conjugative transfer and PhiC31 site-specific recombination. The plate-activated Streptomyces albus J1074 recombinant strain was inoculated into 30 mL of trypticase soy medium (TSB medium), cultivated at 30° C. and 220 rpm for 72 h, and then inoculated with 1% of the inoculum into 3 L of fermentation medium (4 % glucose, 1% glycerin, 3% soluble starch, 1.5% soybean peptone, 1% beef extract, 0.65% peptone, 0.05% yeast extract, 0.1% magnesium sulfate heptahydrate, 0.2% sodium chloride and 0.24% calcium carbonate) In a 5L fermenter (Shanghai Bailun Biotechnology Co., Ltd.), set the stirrer speed at 500rpm, ferment at 30°C for 10 days, and feed 36mL 500g L every day through a peristalt...
Embodiment 3
[0101] Example 3. Using RedEx technology to seamlessly knock out the spnK gene on the spinosyn gene cluster
[0102] After inactivation of the spnK gene (GenBank ID: AY007564.1) of the spinosyn gene cluster, the Saccharopolyspora spinosa mutant can synthesize spinosyn J and spinosyn L (3'-oxo-demethyl spinosyn A / D), spinosyn J and spinosyn L are important raw materials for the synthesis of spinetoram.
[0103] 1. Preparation of the ampccdB traceless modified gene cassette carrying a 20bp terminal homology arm:
[0104] Use the p15A-ccdB-amp plasmid as a template for PCR amplification to obtain the ampccdB traceless modified gene cassette. The primers for PCR amplification are as follows, and the underline is the PacI restriction site:
[0105] delKampccdB-1: 5'-TTGAGCAGGTCCAGGTACAGCGCGTTCTGGGAGGGCATGTCAATTCCTCCTCAGCCGCCCTCGACGCCGA TTAATTAA TTTGTTTATTTTTCTAAATAC-3',
[0106] delKampccdB-2: 5'-CCGCGCCGGGGTTCGTGCCCCGGCAAGCGCTCGGCGTCGAGGGCGGCTGA T TAATTAA TTTGTTCAAAAAAAAGCC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com