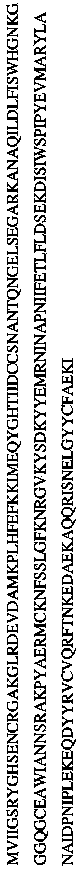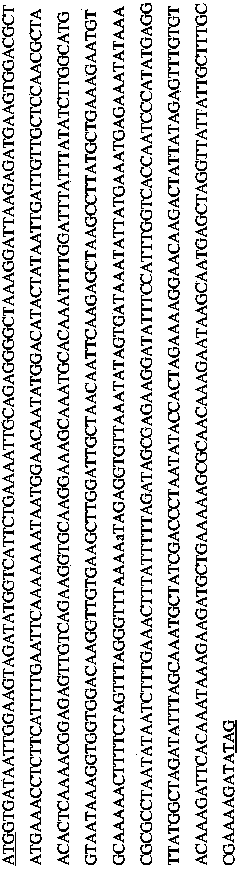Bacteriophage lytic enzymes as alternative antimicrobials
一种噬菌体裂解酶、抗微生物的技术,应用在作为可替代的抗微生物剂的噬菌体裂解酶领域
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0147] The following example shows phage lytic isolation of Clostridium perfringens.
[0148] Bacterial hosts, phage isolation and propagation
[0149] Clostridium perfringens isolates 39 and 26 were used as strains obtained from 0.45 μm filtered offal wash (O) or chicken manure (F) at a chicken processing facility in Athens, GA. Host for phage proliferation. Host strains were characterized by standard methods and 16S sequence analysis (Collins et al., 1994; Wise and Siragusa, 2005). Initial screens for phage lysis of Clostridium perfringens were performed on filtered samples obtained from poultry (gut material), soil, sewage and poultry processing effluent water. Bacterial viruses capable of lysing strains of Clostridium perfringens were identified by sampling and titration of isolated phage-susceptible strains. Clostridium perfringens was propagated by standard procedures (Ackermann and Nguyen, 1983) using anaerobic chamber-grown C. perfringens isolates 39 and 26 ho...
Embodiment 2
[0152] The following examples illustrate molecular cloning, sequencing, annotation and phylogenetic analysis of genomic DNA.
[0153] Following purification of phage genomic DNA, nucleic acids were read spectrophotometrically and restriction enzyme digested at 260 / 280 nm, followed by agarose gel electrophoresis to assess purity (Sambrook et al., supra). Full-length genome sequencing of the phage genome was performed by MWG Biotech, Inc High Point, NC USA. Briefly, phage genomic DNA was sheared, blunt-ended and dephosphorylated using a sprayer (Sambrook et al., 1989). After transformation, blunt-end ligate DNA fragments of the desired size (1 to 4 kb) into pSmart (Lucigen TM ) for the proliferation of Escherichia coli. Clones were sequenced such that approximately 14-fold redundancy was obtained for the genome including primer walking to fill gaps (Fouts et al., 2006). restriction enzyme Hin dIII, Eco RI, Eco RV, Alu I and cla I perform molecular cloning to cut the...
Embodiment 3
[0156] The following example shows virion purification, two-dimensional electrophoresis and proteomic analysis of isolated CP39O (PTA-11081) and CP26F (PTA-11080) phage.
[0157] Purified virion sample preparation for two-dimensional (2D) gel electrophoresis. Phage protein purification was accomplished by adding four volumes of cold (-20°C) acetone to pellet the centrifuged phage for at least one hour. Centrifuge the soluble protein fraction in acetone at 16,000 x g for 10 min at 4 °C. The pellet was washed three times in cold acetone / water (4:1 ), then centrifuged, and dried under vacuum (Champion et al., 2001; Lee and Lee, 2003). To ensure reproducible gel electrophoresis, gel electrophoresis was performed at 37°C according to the manufacturer's instructions (New England BioLab TM ) The purified virion protein fraction was digested with N-glycosidase F (PNGase F) for 1 hr to cleave oligosaccharides from N-linked glycoproteins (Maley et al., 1989) prior to extraction with ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com