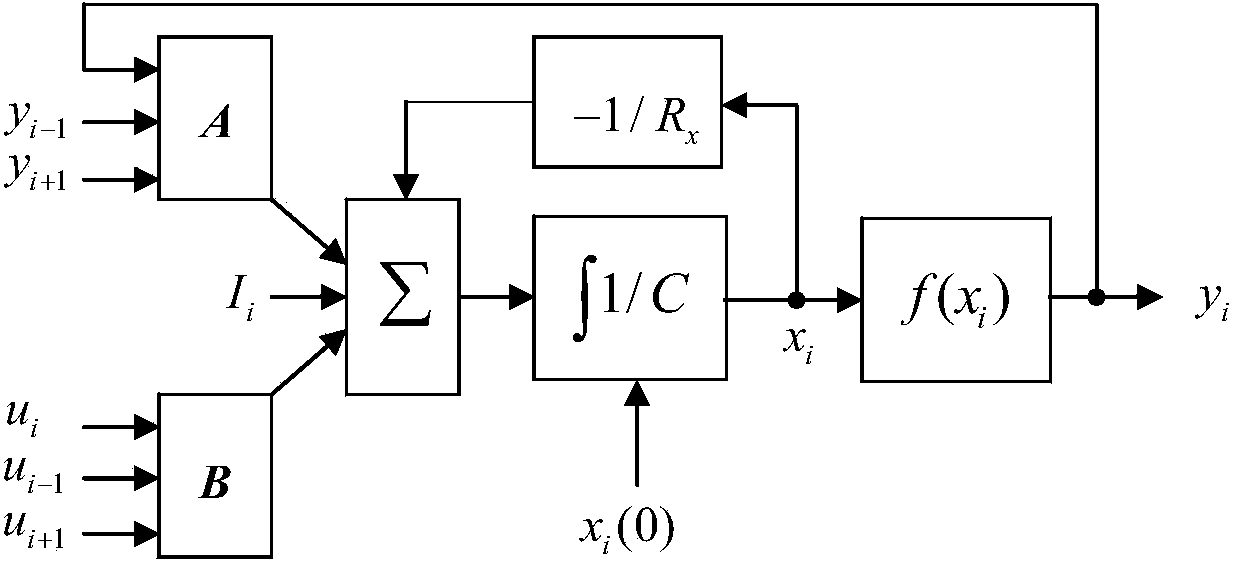
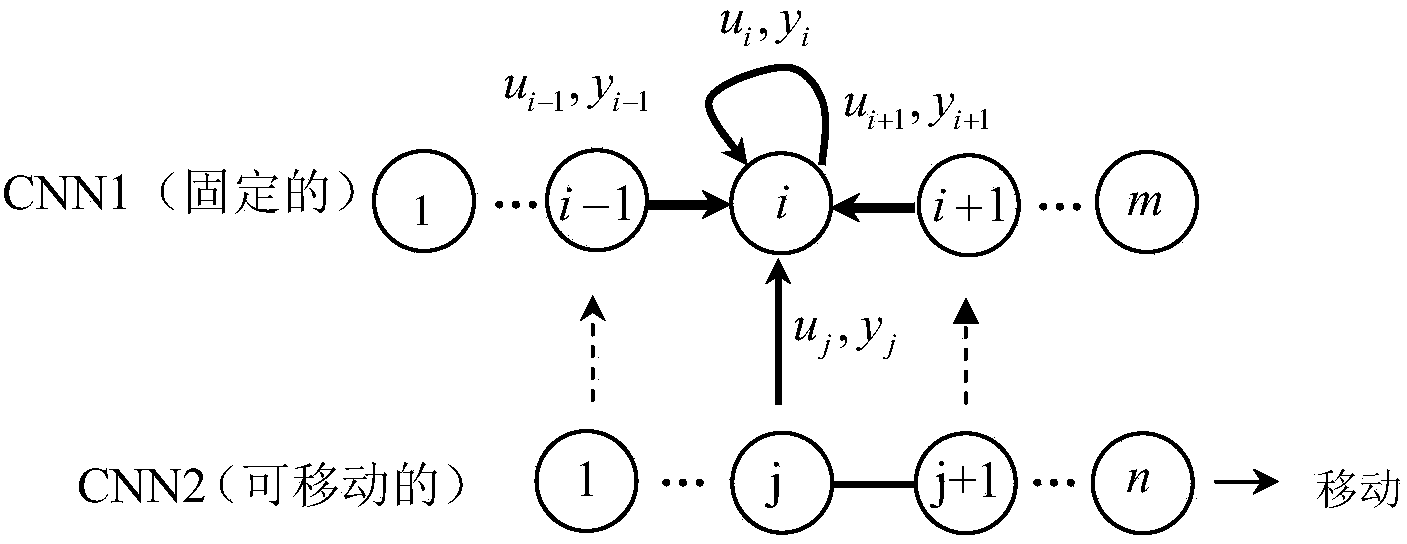
Method for detecting DNA sequence similarity by using one-dimensional cell neural network
A technology of DNA sequence and neural network, applied in the field of DNA sequence similarity detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0073] The implementation process of the present invention will be further specifically described below in conjunction with two DNA base sequences that are actually taken from the NCBI database.
[0074] The selected DNA base sequence identification numbers are S62051 and NM_008134 respectively. For the convenience of display, only two sequence fragments are selected to illustrate the implementation process. The details of these two sequence fragments are shown in Table 1:
[0075] DNA base sequence
Sequence base fragment code
S 1
8
AAGCTCTG
S 2
6
CAGCAT
[0076] Table 1
[0077]Two DNA sequence fragments as shown in Table 1 S 1 and S 2 , and the number of bases they contain is K 1 =8 and K 2 =6, according to the 4 sub-steps under step 3, pair S 1 and S 2 Align as follows:
[0078] ① According to step 3.1 of step (3), initialize the designed one-dimensional dual cellular neural network:
[0079] m=8+1=9...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com