Genetic modification method of target gene in genome
A genetic transformation and genome technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, and the use of vectors to introduce foreign genetic materials, can solve the problem of low probability of success
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
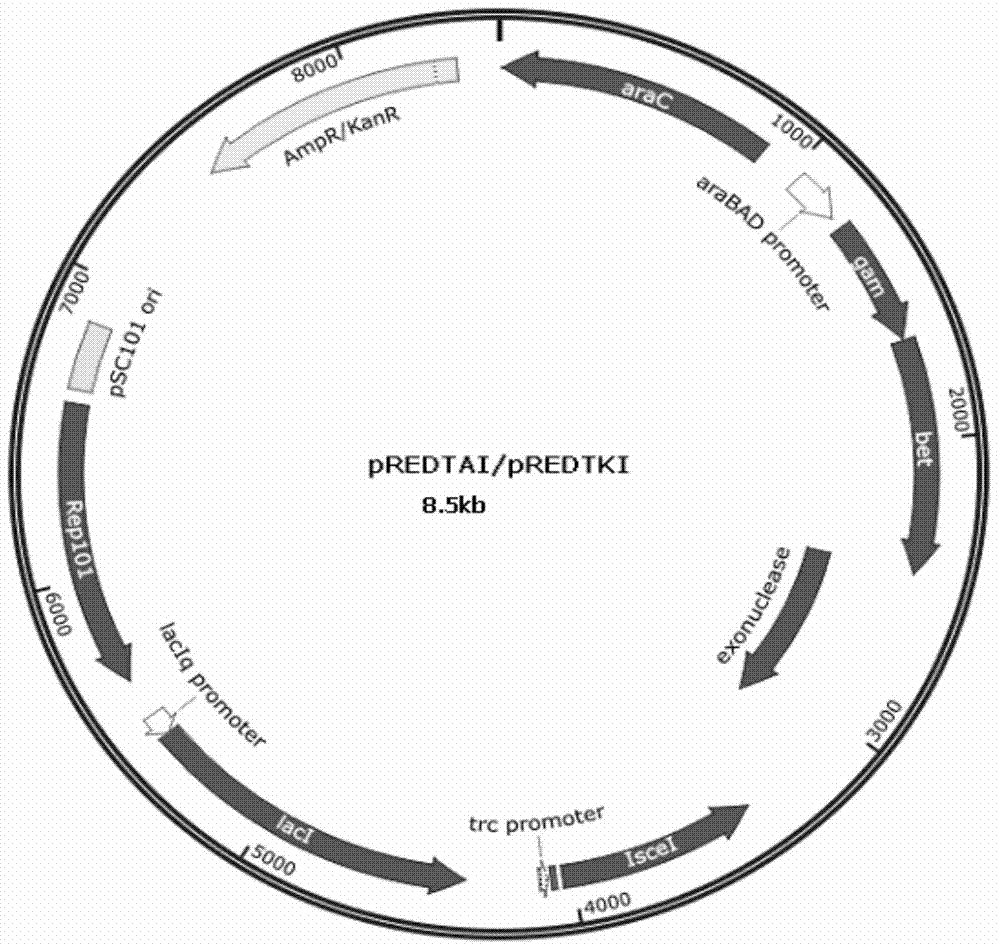
[0185] Embodiment 1, construction Red recombinase+I-SceI endonuclease bifunctional auxiliary (helper) plasmid
[0186] 1. Construction of plasmids expressing I-SceI with rha promoter and λRED recombinase with arabinose promoter
[0187] The specific operation steps are: rhaRas (NcoI) and rhaov primers amplify the genome of Escherichia coli w3110 to obtain Fragment B (about 2 kb) containing a rhamnose promoter (rhaB promoter). I-SceIup and I-SceIdn (NcoI) amplify the pUC19RP12 plasmid (Nucl. Acids Res. (1999) 27 (22): 4409-4415) to obtain fragment C (about 0.7kb) containing the I-SceI endonuclease gene .
[0188] rhaRas (NcoI): ACCATGGGGCATGGCGAATTAATCT (SEQ ID NO: 1);
[0189] I-SceIdn (NcoI): TCCATGGTTATTATTTCAGGAAA (SEQ ID NO: 2);
[0190] I-SceIup: ATGCATCAAAAAAACC (SEQ ID NO: 3);
[0191] Rhaov: GGTTCATTACCTGGTTTTTTTTGATGCATAATGTGATCCTGCTGAAT (SEQ ID NO: 4).
[0192] The two fragments B and C were linked together by Overlap PCR and cloned into pMD18simple-T vector. T...
Embodiment 2
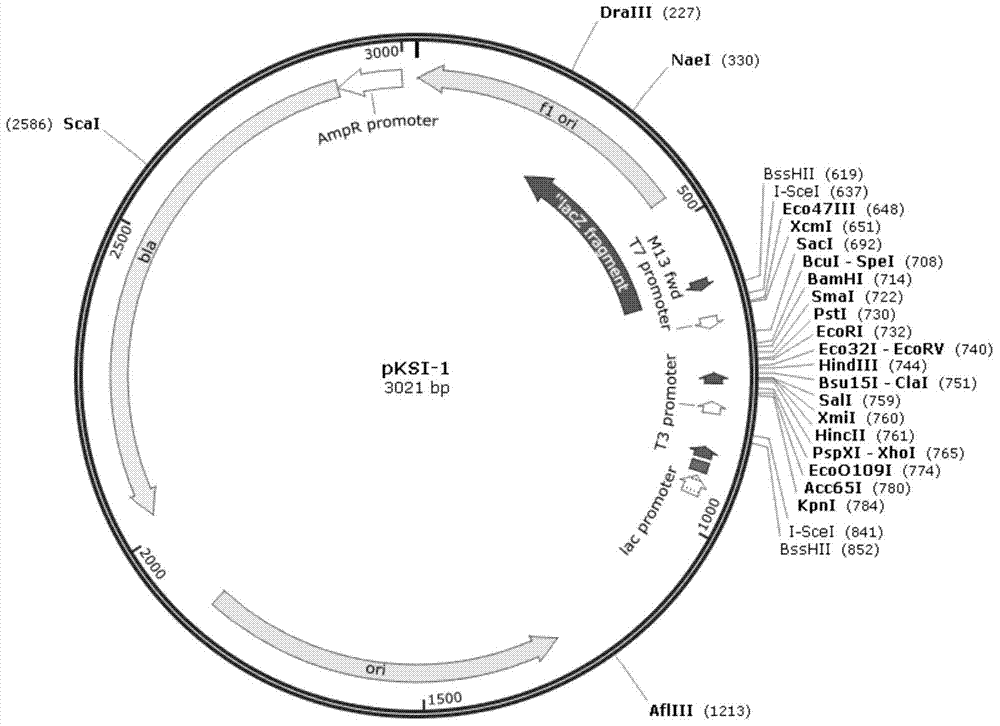
[0215] Embodiment 2, construction has the cloning vector of I-SceI restriction site
[0216]First, a 1.5 kb fragment containing the kanMX gene was excised from pUG6 (from EUROSCARF; see Güldener, U. et al.; 1996; Nucleic Acids Research24, 2519-2524) with NotI, and connected into pBluecript II KS(-) (Nucleic Acids Res17 :9494, from the NotI site of Stratagene) to obtain pKS-K.
[0217] The kanMX gene of pKS-K was amplified with primers T3_IsceI and T7_IsceI, digested with BssHII, and reconnected into the BssHII site of pBluecriptIIKS(-). According to the connection direction, two plasmids, pKSKI-1 (the direction of the kanMX gene and the direction of the lacZ fragment of pBluecriptIIKS(-) itself are opposite) and pKSKI-2 (the direction of the kanMX gene and the direction of the lacZ fragment of pBluecriptIIKS(-) itself are the same) were obtained respectively.
[0218] T3_IsceI: AAGCGCGC CCAATTAACCCTCACTAA (SEQ ID NO: 13);
[0219] T7_IsceI: CAGCGCGC TGTAATACGACTCACTA...
Embodiment 3
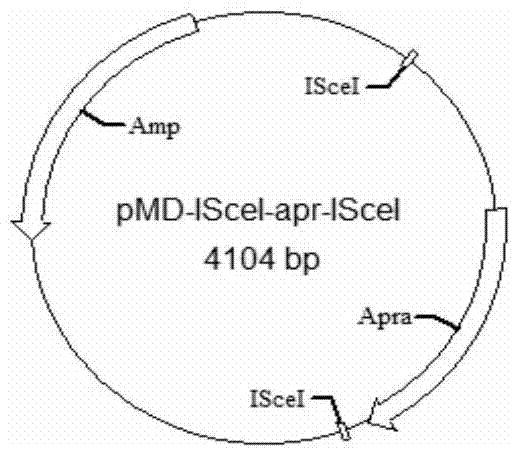
[0227] Embodiment 3, construction is used for knocking in the template plasmid
[0228] Using pIJ773 and pIJ778 plasmids as templates (PNAS2003100 (4) 1541-1546) respectively, the primer pair Apr-ISceI-F and Apr-ISceI-R were amplified to obtain 1.5 kb containing apramycin (Apr) or spectinomycin ( Spc) resistant fragments. After adding A, it was connected into pMD18-T simple (TAKARA) to obtain pMD-ISceI-apr-ISceI and pMD-ISceI-spc-ISceI plasmids. Such as image 3 and Figure 4 .
[0229] Apr-ISceI-F: 5'TCCCCCGGGGCTAGGGATAACAGGGTAATATTCCGGGGATCCGTCGACC3' (SEQ ID NO: 15);
[0230] Apr-ISceI-R: 5'TCCCCCGGGGCATTACCCTGTTATCCCTAGTGTAGGCTGGAGCTGCTTC3' (SEQ ID NO: 16).
[0231] The pKD3 plasmid (PNAS2003100(4)1541-1546) was digested with XbaI to obtain a 1.0 kb fragment containing chloramphenicol (Cm) resistance. At the same time, the pMD-ISceI-apr-ISceI plasmid was also digested with XbaI, the apramycin resistance fragment was excised, and the vector backbone was retained. Conn...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap