Separation method of liver cancer stem cells
A technology for liver cancer stem cells and liver cancer cells is applied in the field of cell separation, which can solve the problems of lack of specific marker identification and separation of LCSCs, and achieve the effects of good application prospects, improved success rate and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1. Isolation of liver cancer stem cells
[0028] To isolate liver cancer stem cells, follow the steps below:
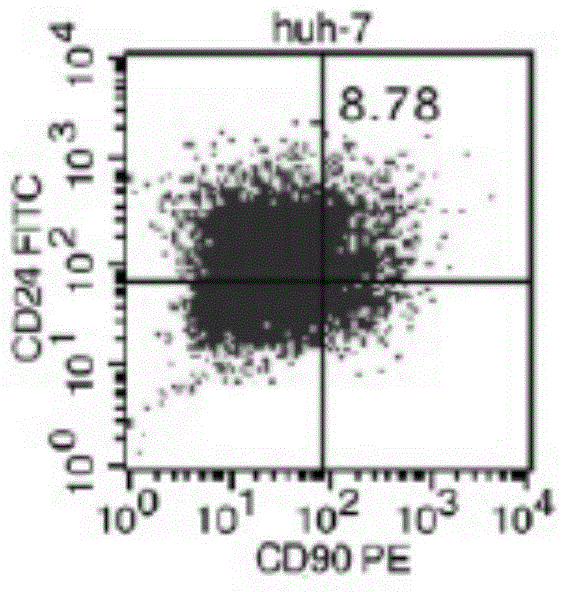
[0029] 1) Liver cancer cell line Huh-7 was cultured with 10% fetal bovine serum DMEM containing 100u / mL penicillin and streptomycin and high glucose at 37°C, 90% relative humidity, 5% CO 2 cultured in an incubator.
[0030] 2) When the cell coverage rate reaches 90%, the hepatoma cell culture solution obtained in step 1 is discarded, washed with PBS, digested with 0.25% trypsin until the cells fall off, the digestion is terminated, and passaged.
[0031] Cell subculture is performed as follows:
[0032] When it is observed that the confluence of cells in the culture flask is 90% (cell growth is in the logarithmic phase), the cells after successful thawing and resurrection are separated and cultured.
[0033] ① Discard the old culture medium and wash 1-2 times with PBS solution (without calcium and magnesium ions).
[0034] ② Add 1ml of digestion sol...
Embodiment 2
[0041] Example 2. Detecting spheroidizing ability of 4 kinds of liver cancer cell subpopulations
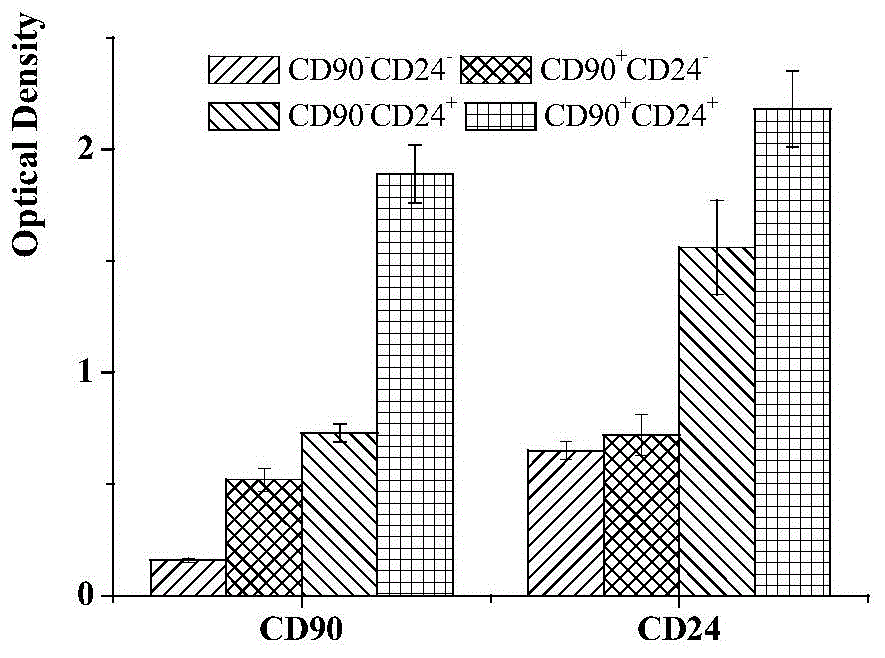
[0042]On the ultra-low adhesion culture plate, the four subgroups of liver cancer cells CD90+CD24+, CD90+CD24-, CD90-CD24+, CD90-CD24- were inoculated into DMEM / F12 medium without serum (containing 20 ng / mL bFGF (basic fibroblast growth factor, cationic polypeptide composed of 155 amino acids), 20ng / mL EGF (oligopeptide-1, active polypeptide composed of 53 amino groups), 20ng / mL LIF (leukemia inhibitory factor) , 2% B27 (cell culture additive), 100u / mL of penicillin and 100u / mL of streptomycin), observe the number of spheres of cells in 7 days, 14 days, 21 days and 28 days, and carry out the selected cells After culture and expansion, the cells were collected at the 3rd, 5th, 7th, 10th, and 13th passages, and the ability to form spheres was repeatedly verified three times. Using t statistical analysis, it was found that CD90+CD24+ liver cancer cells were significantly different f...
Embodiment 3
[0043] Example 3. Detection of clonal formation rate of 4 kinds of liver cancer cell subpopulations
[0044] The 4 kinds of liver cancer cells were inoculated in 6-well plates with 1000 cells per well, cultured in low-serum culture medium made of 5% FBS (fetal bovine serum) and double antibodies, the medium was changed every 2 days, and terminated after 3 weeks Culture, discard the supernatant, wash twice with PBS, fix with methanol, add Giemsa stain reagent 1mL / well, stain for 30min, then slowly wash away the staining solution with running water, and air dry. Finally, count the colonies with more than 50 cells under an ordinary light microscope, and calculate the colony formation rate (PE, PE=number of colonies / number of seeded cells×100%), CD90 + CD24 + The colony formation rate of liver cancer cells was (53.31±0.85)%, much higher than that of CD90 + CD24 - , CD90 - CD24 + , CD90 - CD24 - , with significant difference (P<0.01, P<0.001, P<0.001).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com