Crispr/Cas9-induced scale-missing zebra fish mode and establishment method
A technology of zebrafish and scales, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
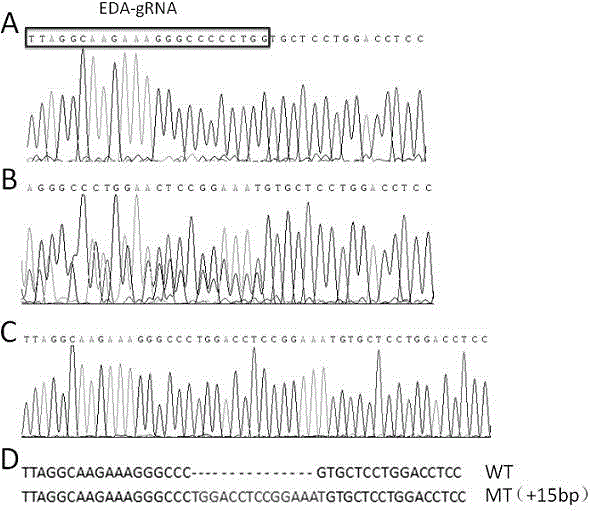
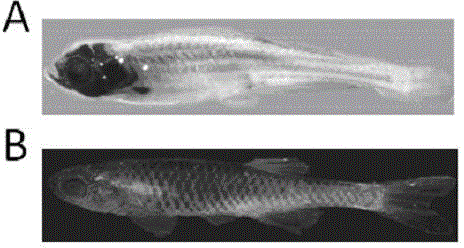
[0037] Example 1 A scale-deleted zebrafish model induced by Crispr / Cas9, which refers to a scale-deleted zebrafish that contains a base insertion at the exon 4 site of the EDA gene.
[0038] The establishment method of the scale-deficient zebrafish model comprises the following steps:
[0039] (1) Using the Ensemble online database, search for the zebrafish EDA gene sequence no: ENSDARG00000074591 and download it, and search for the PAM (protospacer-adjacent motif) sequence in this sequence, namely 5'-GGNNNNNNNNNNNNNNNNNNNNNGG-3', the targeting effect found in the exons of the EDA gene The sequence is TTAGGCAAGAAAGGGCCCCC [TGG], and the mutation detection primer F-eda: ttgttttgcttctcatcagttg and the mutation detection primer R-eda: tttgctctgctgcttcactc are designed and designed.
[0040] (2) Sequencing verification of the wild-type allele of the target sequence:
[0041] Five 24hpf wild-type zebrafish embryos were randomly selected, the genomic DNA template was extracte...
Embodiment 2
[0071] Example 2 The scale-deleted zebrafish model was applied to the verification of the function of skin attachment-related genes and the screening of drugs for ectodermal dysplasia.
[0072] PAM sequence
[0073] 5'-GGNNNNNNNNNNNNNNNNNNNGG-3'
[0074]
[0075] targeting sequence
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com