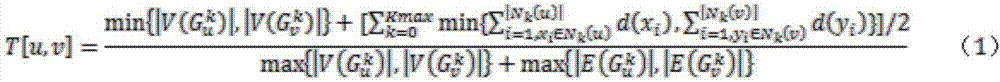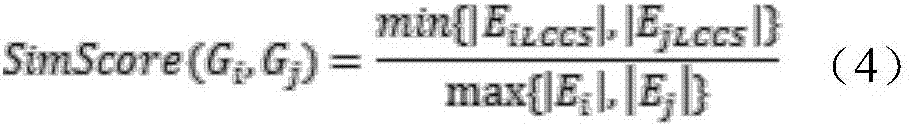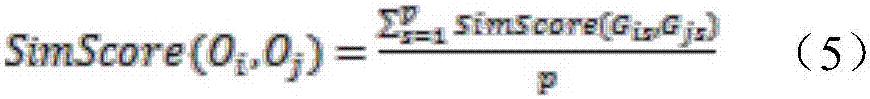Method for rebuilding species system generation tree based on alignment of multiple metabolic pathways
A phylogenetic tree and path technology, applied in biological systems, evolutionary organisms, bioinformatics, etc., can solve problems such as limited node mapping information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0040] The method of reconstructing the phylogenetic tree of species based on the comparison of multiple metabolic pathways, the specific steps are as follows:
[0041] 1) Establishment of multiple metabolic pathway composite maps:
[0042] 1.1) Calculation of node similarity:
[0043] For the metabolic pathway P, let G p =(V p ,E p ) represents the metabolic pathway P, where G p is a directed graph, V p is G p The vertex set of E p is G p The set of directed edges, G p Vertex u in i and u j Denotes the reaction r in P i and r j . if r i One of the output compounds is r j an input compound of , then u i and u j There exists a line from r i to r j The directed edge, if r i , r j are reversible, then there is also a path from r j to r i The directed edge of .
[0044] k is a positive integer, for graph G p For any node u in , define the set of k neighbors of u: N k (u),N k (u) is V p A set of nodes for which u does not belong to N k (u) and for any x...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com