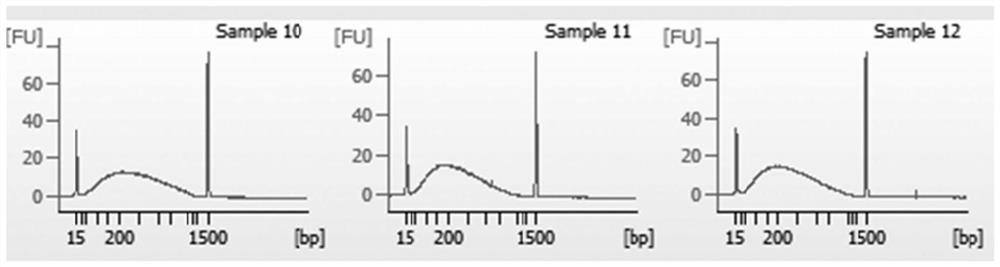
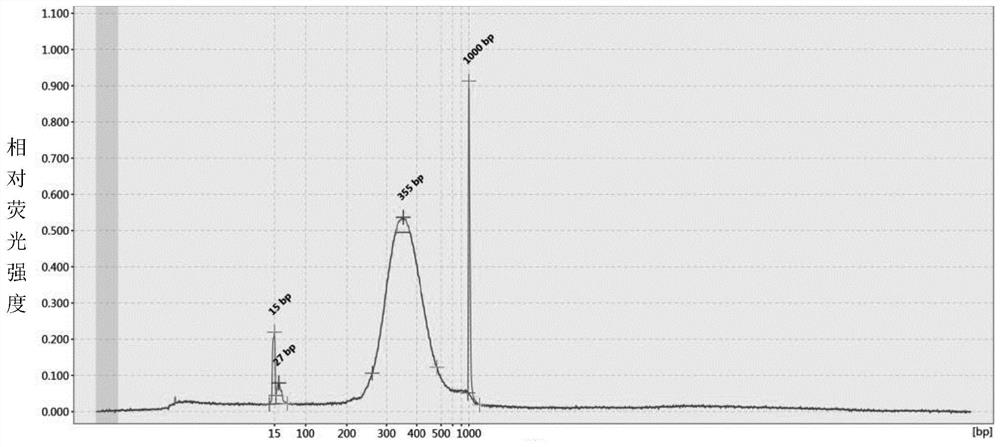
Preparation method of quantitative standard substance for high-throughput sequencing library of illumina platform
A quantitative standard and sequencing library technology, applied in the direction of microbiological-based methods, biochemical equipment and methods, microbiological determination/inspection, etc., to achieve accurate quantification and high method accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Preparation and determination of quantitative reference materials for high-throughput sequencing libraries on the Illumina platform
[0047]1. Materials and methods:
[0048] 1. Escherichia coli (E.coli) genomic DNA extraction
[0049] After streak culture of Escherichia coli strain BL21(DE3), pick a single colony for overnight liquid culture, centrifuge at 5000g, discard the supernatant, collect the bacteria, and use Kangwei Century’s general genome extraction kit (CW2298) for genome extraction .
[0050] When the tested sample is a human genome, it can be replaced with other non-human genomic DNA; when the tested sample is a microorganism, it can be replaced with other non-target microorganisms or human-derived genomic DNA.
[0051] 2. Random interrupt
[0052] The above-mentioned extracted and purified E.coli genomic DNA was randomly interrupted by the Covaris S220 instrument, and the processing conditions were: intensity: 5, time: 6min, sample volume: 100μL, DNA ...
Embodiment 2
[0090] Example 2 High-throughput sequencing library quantitative reference material applicability verification
[0091] 1. Materials and methods:
[0092] 1. Library Quantitative Reference Material
[0093] The 5-level library quantitative reference material was developed by the China Institute of Metrology.
[0094] 2. Library quantification kit
[0095] Library quantification kits were purchased from KAPA and NEB respectively.
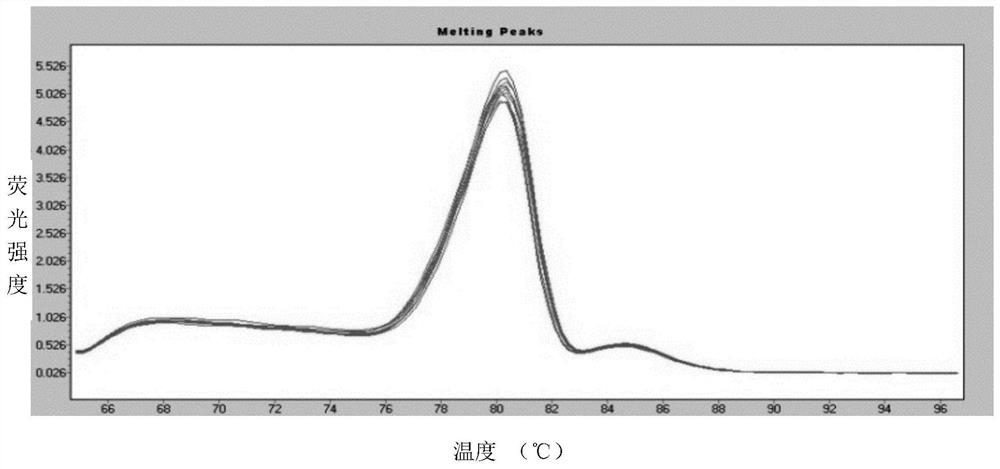
[0096] 3. Measurement of standard substance linearity by fluorescent quantitative PCR
[0097] Roche480 fluorescent quantitative PCR instrument was used to measure the quantitative standards of KAPA Company and Qiagen Company respectively. 20 μL of PCR amplification system: 10 μL of 2×SYBR PCR mastermix, 0.4 μL of bidirectional primer (10 μM), 4 μL of DNA template, ddH 2 O 5.6 μL. PCR reaction program: 95°C for 5min, 95°C for 20s, 60°C for 40s, 45cycles, melting curve: 95°C for 5s, 65°C for 1min, continuous signal acquisition at 95°C.
[0098]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com