Compositions and methods relating to tumor analysis
A tumor, tumor cell technology, applied in the field of compositions and methods related to tumor analysis, which can solve problems such as cancer model limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
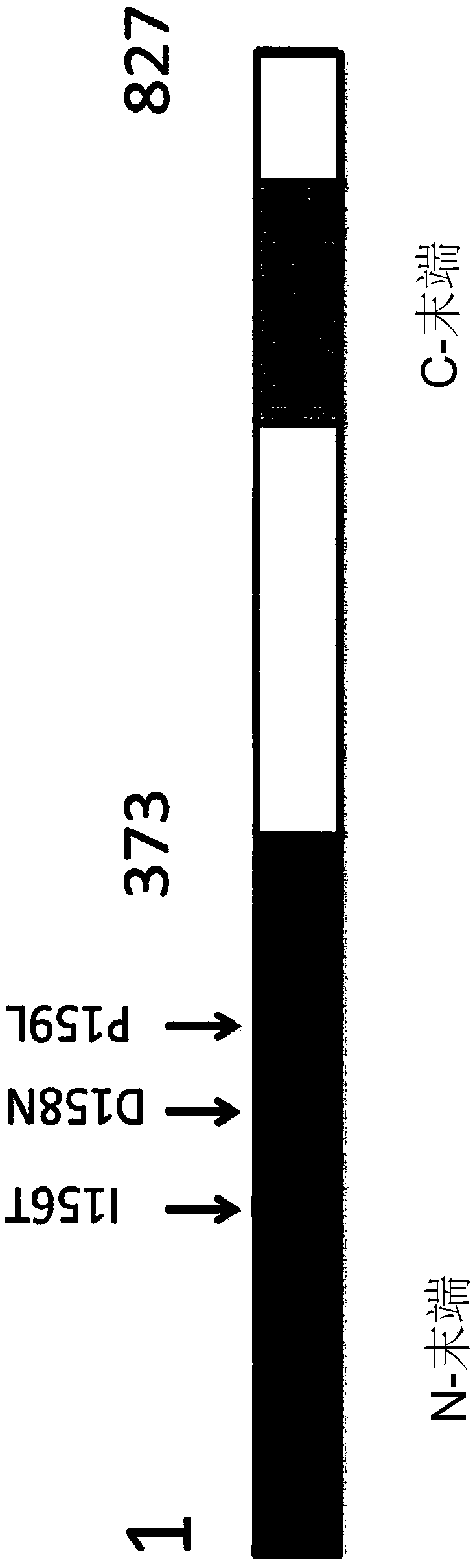
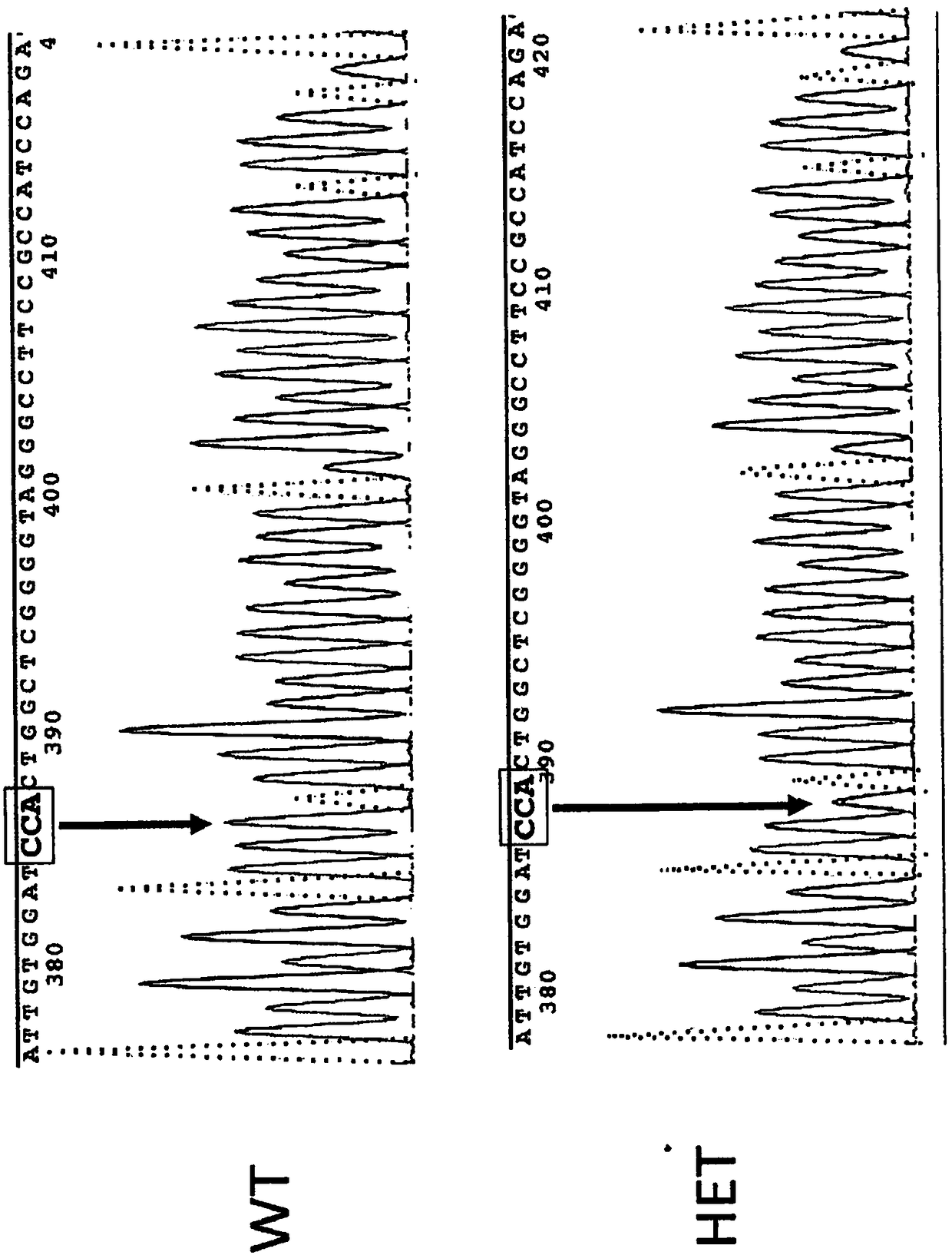
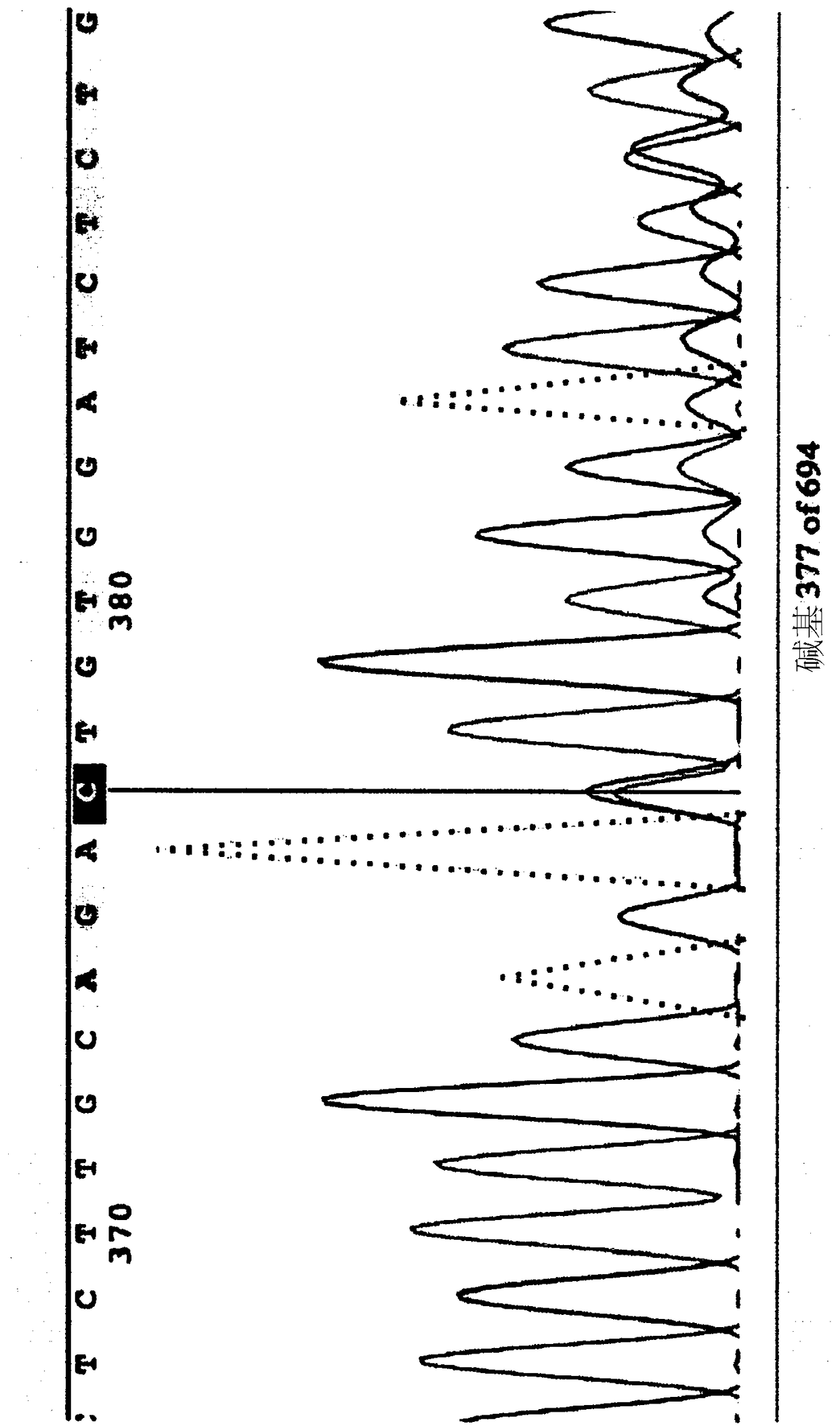
[0178] NSG-BALDP159L mice carrying a point mutation p.P159L in the Rhbdf2 gene were generated using CRISPR / Cas9 technology. Microinjection of Cas9 mRNA, truncated guide RNA (sgRNA), and single-stranded oligonucleotide DNA (ssDNA:
[0179] ) to target the Rhbdf2 locus in NSG embryos to mutate the proline at amino acid 159 of SEQ ID NO:1 to leucine. The underlined codon CTA is a mutated codon introduced into the genome. figure 1 A schematic diagram of mouse iRhom2 (SEQ ID NO: 1 ) is shown.
[0180] sgRNA used to generate mouse (17mer): G CAG ATT GTG GAT CCAC (SEQ ID NO: 7)
[0181] CRISPR / Cas9 protocol:
[0182] 1. Turn on the PCR instrument and centrifuge - close the lid and set the temperature to 4°C.
[0183] 2. RNAase-Zap (working area).
[0184] 3. Remove the following from -80°C to ice (in a labeled box directly in front of the bottom rack)
[0185] a.sgRNA
[0186] b. Cas9 mRNA
[0187] 4. Centrifuge guide RNA and Cas9 mRNA at 20,000x g for 15 minutes at 4°C
...
Embodiment 2
[0235] animal
[0236] Six to eight week old NSG-BALDP159L (n=5) and NSG female mice (n=4) were used to examine tumor growth. Mice were bred and maintained under specific pathogen free (SPF) conditions at the Jackson Laboratory. Food and acidified water were provided ad libitum.
[0237] Xenogeneic tumor cell preparation and administration
[0238] MDA-MB 231 human breast cancer cells were cultured in RPMI medium and grown at 37 °C. MDA-MB 231 human breast cancer cells (3×10 6 ) were injected subcutaneously into NSG-BALDP159L and NSG mice.
[0239] tumor size
[0240] The subcutaneous xenograft tumor diameter was measured daily using an external caliper. To determine tumor volume by external calipers, the maximum longitudinal diameter (length) and the maximum transverse diameter (width) were measured using external calipers. Then the tumor volume is calculated as: 1 / 2 (length × width 2 ) = tumor volume. At the end of the study, tumors were collected and subjected to h...
Embodiment 3
[0249] Such as comparing SEQ ID NO:1 and 3 Figure 6 As shown, mouse iRhom1 and mouse iRhom2 are highly related proteins. Given the high structural identity of mouse iRhom1 and mouse iRhom2, studies were performed to determine whether the functional effects of modifications in iRhoml were similar to those observed for iRhom2 modifications.
[0250] generated Rhbdf1 knockout C57BL / 6 mice, generated iRhom1-deficient mice, and found that iRhom1 deficiency resulted in weight loss. Figure 7 showed a normal-sized Rhbdf1 gene deletion heterozygous (Rhbdf1+ / - ) mice (right) homozygous for the deletion of the Rhbdf1 gene (Rhbdf1 - / - ) size differences between mice (left).
[0251] Rhbdf1 knockout C57BL / 6 mice died at 3–4 weeks of age, as Figure 8 shown in Fig. Rhbdf1 gene deletion heterozygous (Rhbdf1 + / - ) mice showed a normal survival percentage ( Figure 8 upper line in the middle panel), and are viable and fertile, whereas Rhbdf1 gene deletion homozygous (Rhbdf1 - / - ) mice...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com