Prostate cancer detection probe sequence and application thereof
A prostate cancer and sequence technology, which is applied in the field of prostate cancer detection probe sequences, can solve problems such as the inability to guarantee specificity, the influence of quantification limit and detection limit sensitivity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1: The kit for detecting prostate cancer of the present invention
[0046] 1. Probe sequence
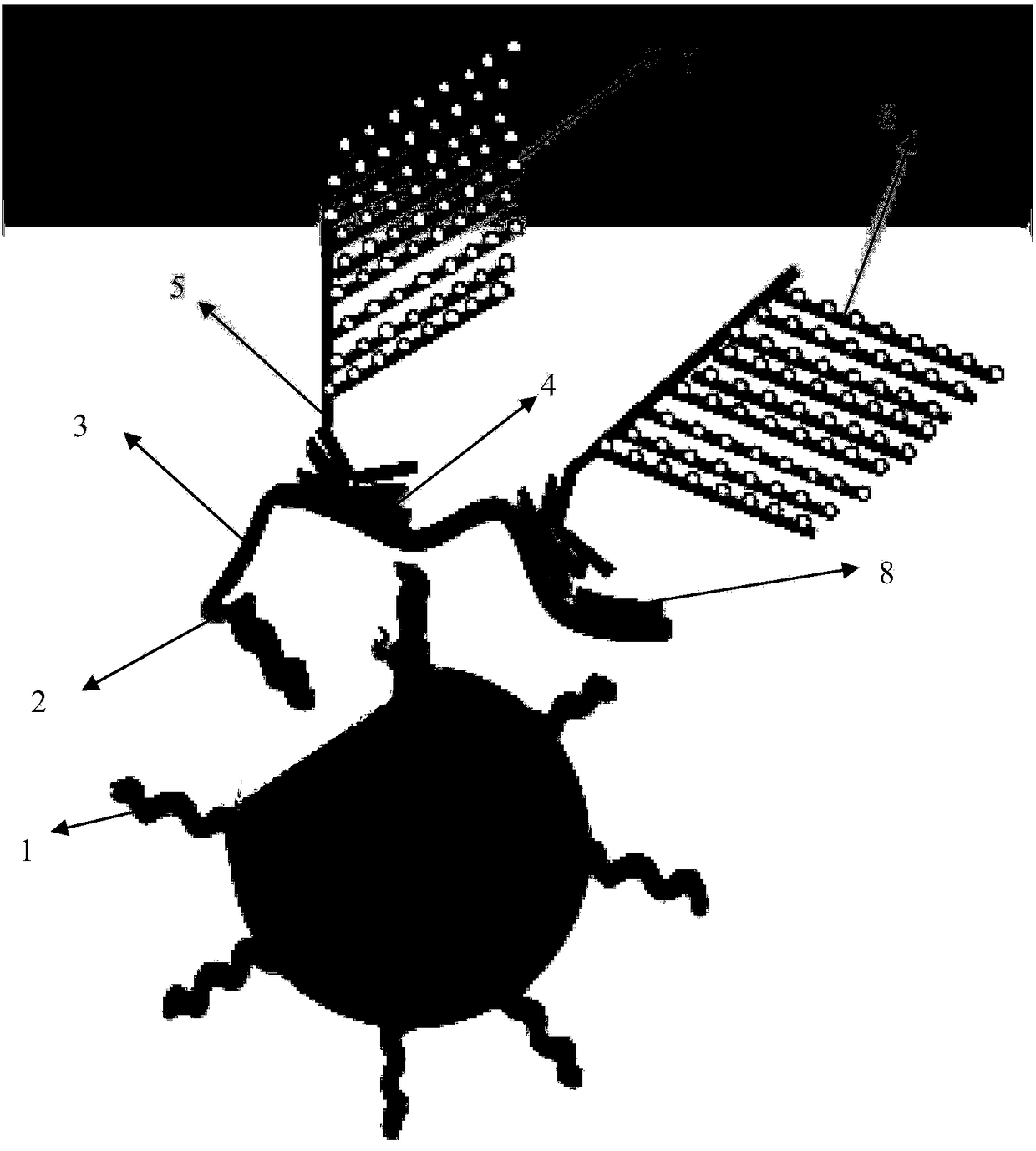
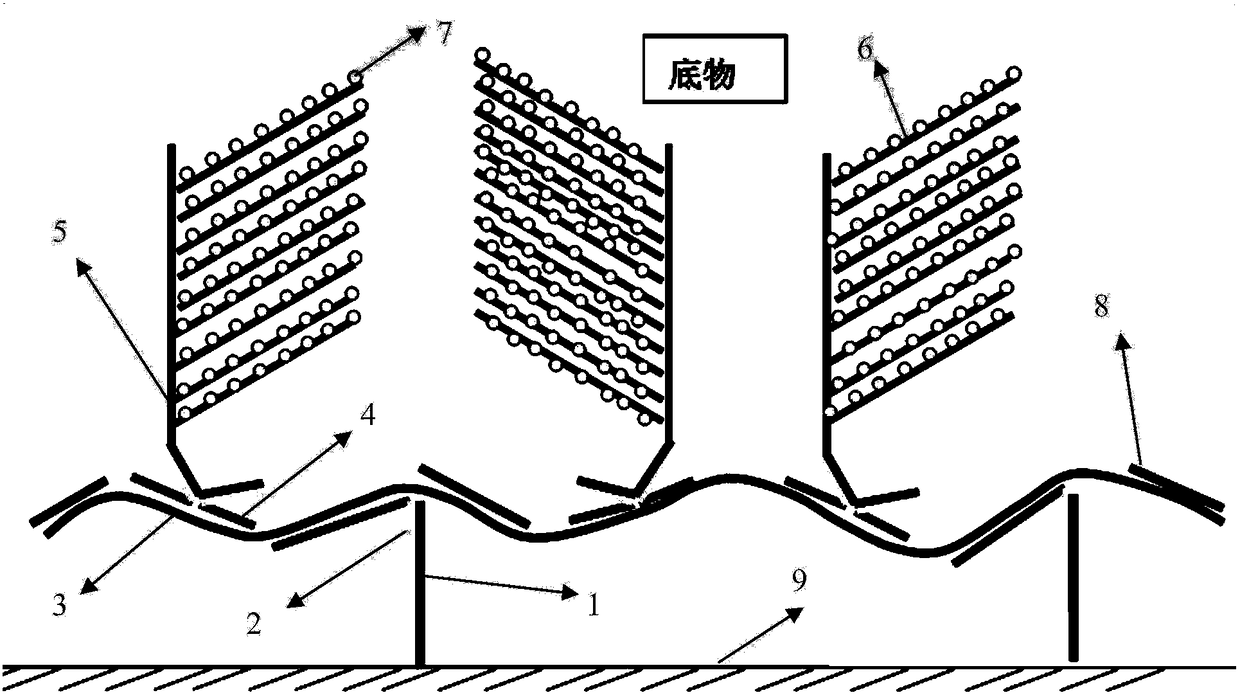
[0047] The capture extension probe sequence is shown in SEQ ID No. 1-14 and SEQ ID No. 78-85, and the K-type labeled extension probe is shown in SEQ ID No. 15-64 and SEQ ID No. 86-131, closed The probes are shown in SEQ ID No. 65-77 and SEQ ID No. 132-148;
[0048] 2. Kit components
[0049] Universal probes coupled to carriers such as magnetic beads or cell culture plates: shown in SEQIDNo.149;
[0050] Signal amplification precursor: ATGCTTTGACTCAG / AAAACGGTAACTTC (amplified precursor pre-leader sequence, two kinds) + AGGATCGTAAGTAACTAAGGACTACC (amplifier pre-leader complementary sequence) + 19 times amplifier pre-leader complementary sequence (SEQ ID No. 150-151 Shown);
[0051] Signal amplifier: GGTAGTCCTTAGTTACTTACGATCCT (amplifier front guide region sequence) + GAATCCATTGAATCCTGTGATT (reporter probe repeat region complementary sequence) + 19 times reporter probe repeat reg...
Embodiment 2
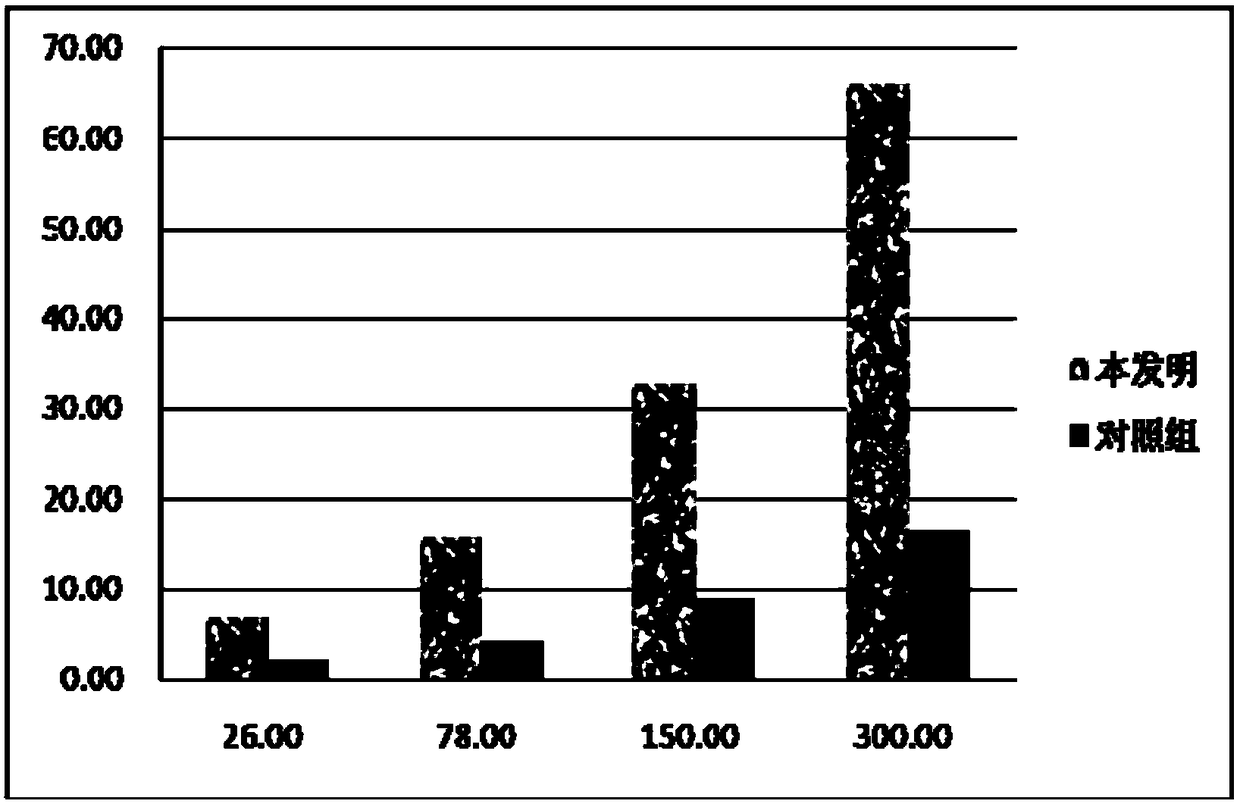
[0054] Example 2: Specificity comparison test
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com