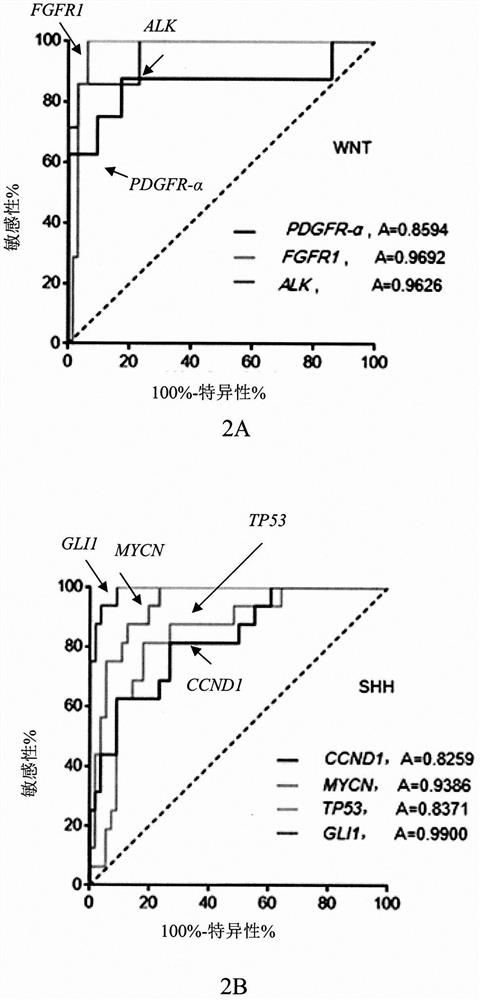
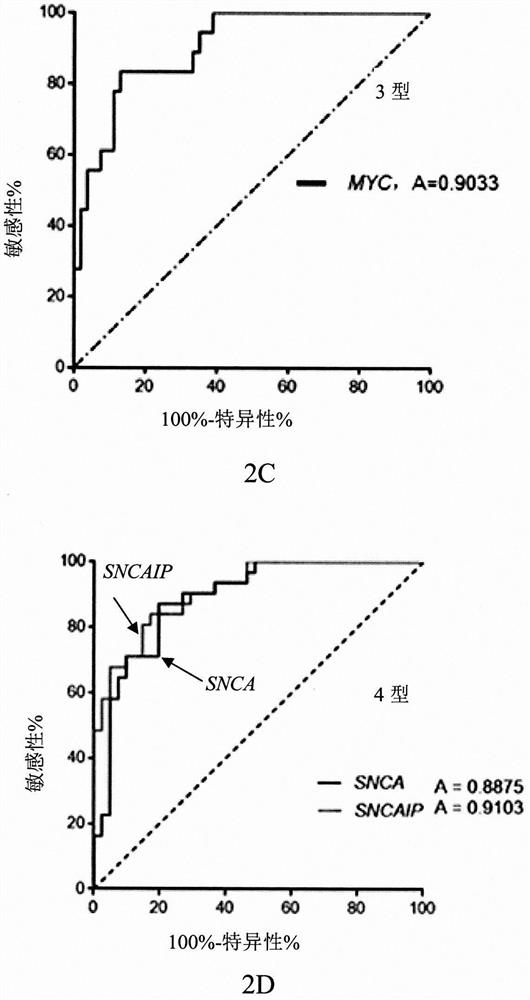
Gene clusters for molecular typing of medulloblastoma and use of the snca gene as a biomarker for medulloblastoma type 4
A technology for medulloblastoma and molecular typing, which is used in the determination/examination of microorganisms, genetic engineering, plant genetic improvement, etc. No further questions such as potential molecular targets for gene-targeted therapy are provided
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1 Classification of Medulloblastoma and Screening of Related Marker Genes of Specific Subtypes
[0063] The FFPE samples of 12 medulloblastoma patients were randomly obtained, and the RNA was extracted with a commercially available kit and the RNA concentration was determined.
[0064] Using the nanoString nCounter technology platform (internal standard genes are CLTC, GAPDH and TUBB), the extracted RNA was detected for the following genes:
[0065] Table 3. Candidate genes
[0066]
[0067]
[0068] Select 12 genes (3 for each subtype) from the 22 recognized molecular subtype-specific genes, and conduct preliminary judgment of MB molecular subtype by Nanostring detection. On this basis, further detect the above 89 genes in MB. The expression of MB molecular subtypes is screened out, and the genes related to MB molecular subtypes that may become molecular therapeutic targets are selected, and other technical methods (DNA next-generation sequencing, gen...
Embodiment 2
[0069] Example 2 NanoString typing and immunohistochemical staining of formaldehyde-fixed paraffin-embedded samples
[0070] Using the 11 tumor-associated genes screened in Example 1 and the known 22 genes associated with MB molecular subtypes (WIF1, TNC, GAD1, DKK2, EMX2, PDLIM3, EYA1, HHIP, ATOH1, SFRP1, IMPG2, GABRA5, EGFL11, NRL, MAB21L2, NPR3, KCNA1, EOMES, KHDRBS2, RBM24, UNC5D, OAS1) constitute a gene group of 33 genes, and the gene group of 22 genes is known to carry out MB sample analysis based on the Nano-String nCounter analysis platform For type comparison, the nanoString nCounter detection probes for each gene are shown in Table 2 above.
[0071] Formaldehyde-fixed paraffin-embedded samples (FFPE samples) were obtained from 106 medulloblastoma (MB) patients from the Third Hospital of Peking University, Beijing Tiantan Hospital, Beijing Sanbo Brain Hospital, and Anhui Provincial Hospital. Total RNA was extracted from slices of 75 MB samples and analyzed based o...
Embodiment 4
[0093] Example 4 Knockout and overexpression of SNCA in MB cells
[0094] Daoy, D283 and D341 cells (from ATCC) were 4 × 10 4 The cell density was seeded in a 6-well plate, and any one of the three SNCA siRNAs (siRNA1, siRNA2, siRNA3) was transfected into the cells using Chemifect transfection reagent (Feng Rui, China) according to the manufacturer's instructions, with a final concentration of 20μmol / L.
[0095] With GV219 ( Figure 4 4A) in ) is the backbone, and insert the SNCA gene fragment through the restriction site XhoI / KpnI using conventional methods in the art to construct the plasmid GV219-SNCA-wt. The constructed GV219-SNCA-wt was used as a template, and the following sequences were used as forward primer and reverse primer respectively for amplification:
[0096] SEQ-F (769-789bp): CGCAAATGGGCGGTAGGCGTG
[0097] SEQ-R (1101-1121bp): CCCACTGTCCTTTCCTAATAA
[0098] The verified sequence contains the SNCA gene sequence ( Figure 4 The italic part of 4B), whic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com