Primer pair sets and kits for detection of genetic modification differences in cytosine deaminase and related molecules in cfDNA
A technology of cytosine deaminase and primer pairs, which is applied in the field of primer pairs and kits for detecting differences in genetic modification of cytosine deaminase and related molecules, and can solve the problems of low mutation frequency, little attention, and peripheral blood difficulties, etc. problem, to achieve good stability and improve sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 Detection of Cytidine Deaminase and Related Molecular Gene Epigenetic Modification Differences in Peripheral Blood Free DNA Primer Pair Set
[0053] Aiming at different regions of cytosine deaminase and related molecular genes, PCR primers were designed and a large number of screening experiments were carried out, and finally a group of detection cytosine deaminase and related genes in peripheral blood free DNA with strong specificity and high sensitivity were obtained. A primer pair set for differences in molecular gene epigenetic modification, the primer pair set consists of the nucleotide sequences shown in SEQ ID No.1 to SEQ ID No.15 in the sequence table;
[0054] Wherein, sequence SEQ ID No.1 and SEQ ID No.2 are respectively the upstream primer and the downstream primer that amplify the AID gene promoter region fragment length is 145bp (AID-P-145), and sequence SEQ ID No.3 is the amplification The probe of this region fragment; SEQ ID No.4 and SEQ ID No....
Embodiment 2
[0090] Example 2 Kit for detecting differences in epigenetic modification of cytosine deaminase and related molecular genes in free peripheral blood DNA
[0091] A kit for detecting differences in epigenetic modification of cytosine deaminase and related molecular genes in free DNA in peripheral blood, said kit comprising the nucleosides shown in the sequence table SEQ ID No.1 to SEQ ID No.15 A primer pair set consisting of acid sequences, PCR reaction solution (Premix Ex Taq TM ), quality control DNA sample and ddH 2 O;
[0092] Wherein, the kit also includes a quality control control DNA sample; the quality control control DNA sample is naked DNA, such as a PCR product that does not have epigenetic modifications; further, it is a better concentration of simulated free DNA , the PCR product needs to be highly diluted to make it equivalent to the sample concentration, and then re-amplified as a template; the quality control control DNA sample includes AID gene promoter regio...
Embodiment 3
[0093] Embodiment 3 establishes detection method
[0094] 1) Extraction of free DNA from peripheral blood;
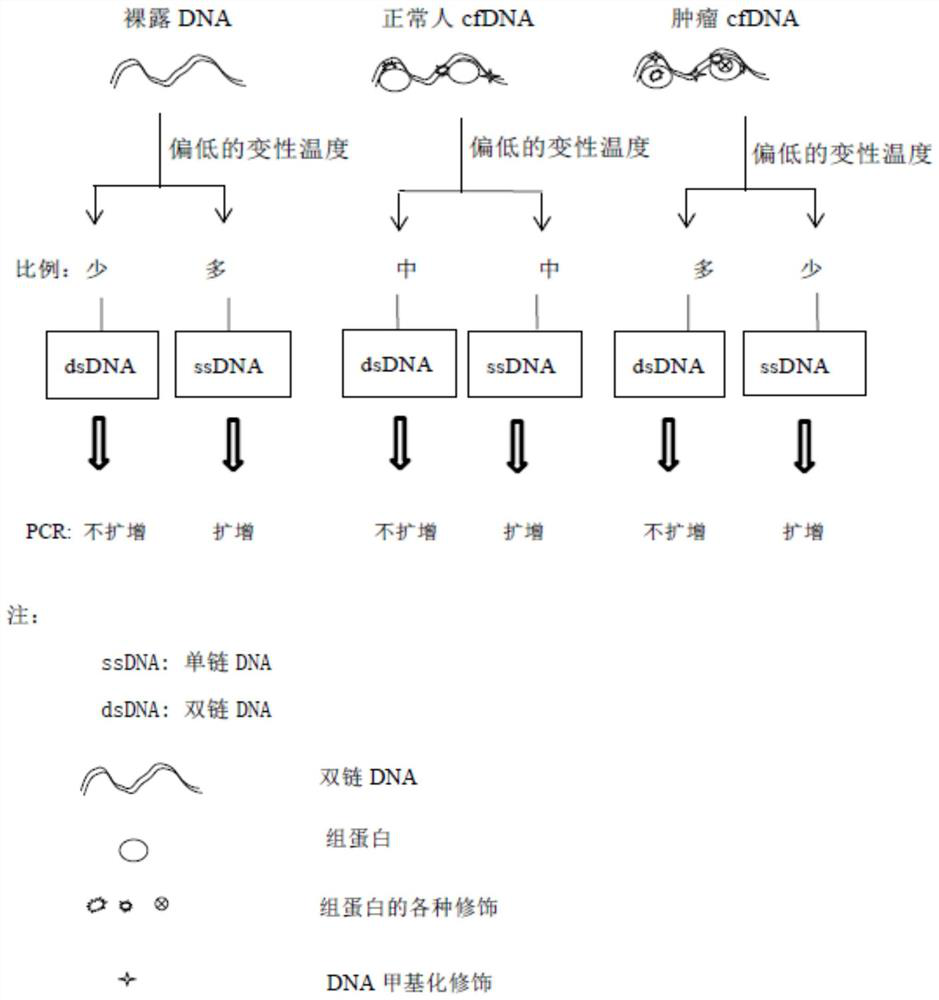
[0095] 2) Using a primer pair group consisting of the nucleotide sequences shown in the sequence table SEQ ID No.1 to SEQ ID No.15, the peripheral blood free DNA and quality control control DNA samples were denatured under high temperature denaturation conditions and low temperature denaturation conditions, respectively. Real-time fluorescent quantitative PCR amplification under the conditions; the qPCR reaction system of each primer set of the reaction body is shown in Table 1-5:
[0096] Table 1 qPCR reaction system of x1 sequence
[0097]
[0098] Table 2 x2 sequence qPCR reaction system
[0099]
[0100] Table 3 x3 sequence qPCR reaction system
[0101]
[0102]Table 4 x4 sequence qPCR reaction system
[0103]
[0104] Table 5 x5 sequence qPCR reaction system
[0105]
[0106] The reaction conditions are:
[0107] High Temperature Denaturation (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com