Internal reference miRNA as esophagus cancer sample and application of internal reference miRNA
A technology for esophageal cancer and samples, which is applied in the field of miRNA detection, can solve the problems of miRNA difficulties, lack of internal reference genes, and limitations of miRNA research and application, and achieve good stability, consistent expression abundance, and high serum expression abundance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] This embodiment aims at preliminary screening to obtain candidate esophageal cancer internal reference miRNAs, and the specific screening methods and results are as follows:
[0033] (1) Select the following seven miRNAs as candidate internal reference miRNAs for esophageal cancer: hsa-miR-361-5p, hsa-miR-186-5p, hsa-miR-26a-5p, hsa-miR-191-5p, hsa- miR-451a, hsa-miR-423-5p and hsa-miR-320a.
[0034] (2) With the help of the data source of TCGA (The Cancer Genome Atlas) database, the expression levels of the aforementioned 7 miRNAs in esophageal cancer tissues and their adjacent tissues were investigated. No significant difference.
[0035] (3) With the help of the data source of the miRNA-seq database, the expression levels of the aforementioned seven miRNAs in serum were investigated, and the results showed that the abundance of hsa-miR-361-5p and hsa-miR-186-5p in serum Low, not suitable as internal reference miRNA.
[0036] (4) According to literature review, hsa...
Embodiment 2
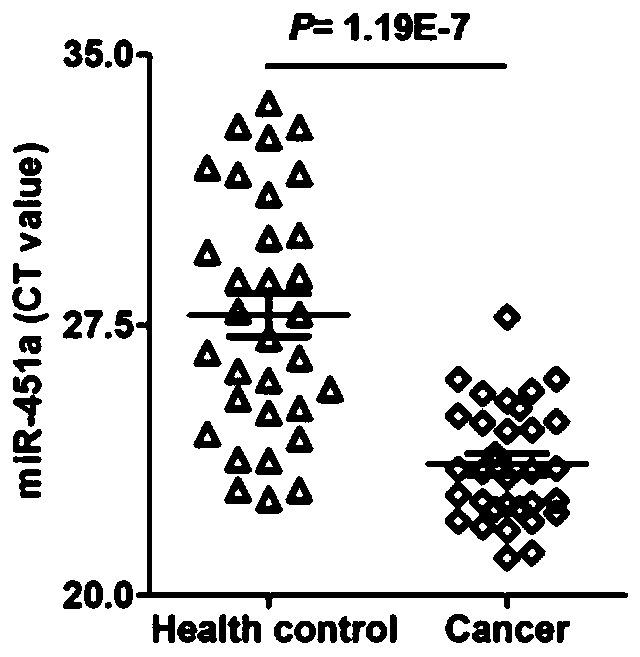
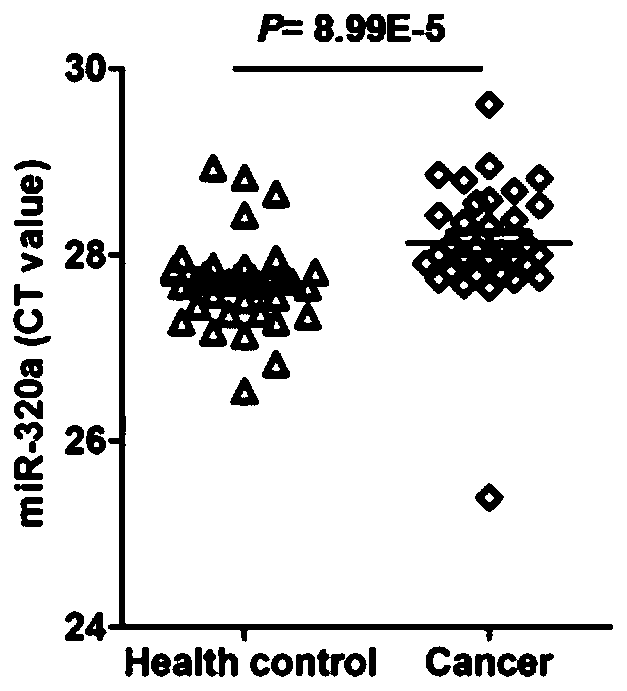
[0039] The purpose of this example is to investigate the expression abundance of hsa-miR-423-5p, hsa-miR-451a and hsa-miR-320a in the samples of normal subjects and esophageal cancer subjects in the Chinese population, in order to investigate their Whether it is suitable as an internal reference miRNA for esophageal cancer. Among them, the subjects consisted of 32 normal subjects and 32 esophageal cancer subjects, all selected from the Chinese population; the test samples were serum, and the detection method was qRT-PCR. The specific detection steps are as follows:
[0040] (1) RNA extraction
[0041] Using miRNeasy Serum / Plasma kit (Qiagen) to extract serum RNA, the operation steps are as follows: 250 μl serum samples were centrifuged (10,000 RPM, 5mins) to remove cell debris, 200 μl supernatant was transferred to a new RNase-removing centrifuge tube; 1,000 μl of The QIAzol lysate was finally dissolved in 30 μl RNase-removed water according to the operation steps provided i...
Embodiment 3
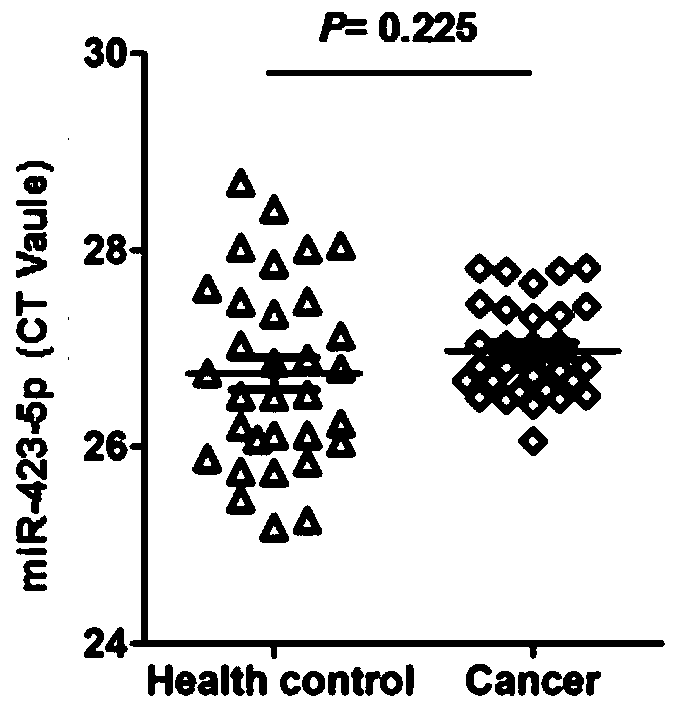
[0059] This example aims to expand the number of samples and further verify the expression level of hsa-miR-423-5p in samples from normal subjects and esophageal cancer subjects. Among them, the subjects consisted of 100 normal subjects and 100 esophageal cancer subjects, all selected from the Chinese population; the test samples were serum, and the detection method was qRT-PCR. Refer to Example 2 for specific detection steps. For specific test results, see Figure 4 .
[0060] according to Figure 4 The results shown show that the expression levels of hsa-miR-423-5p in the serum of normal subjects and esophageal cancer subjects are basically the same without significant difference (P=0.18), and the expression abundance is high, which is suitable as a Internal reference genes of miRNA in esophageal cancer.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com