A kit for detecting methylation at the 5' end of gp5 gene and its detection method
A technology of terminal methylation and kit, which is applied in the field of kits for detecting methylation at the 5' end of GP5 gene, can solve the problems of false positives, expensive instruments, complicated operations, etc., to prevent competition inhibition, high sensitivity, and result analysis. easy effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: Preparation of Lateral Flow Nucleic Acid Detection Test Paper
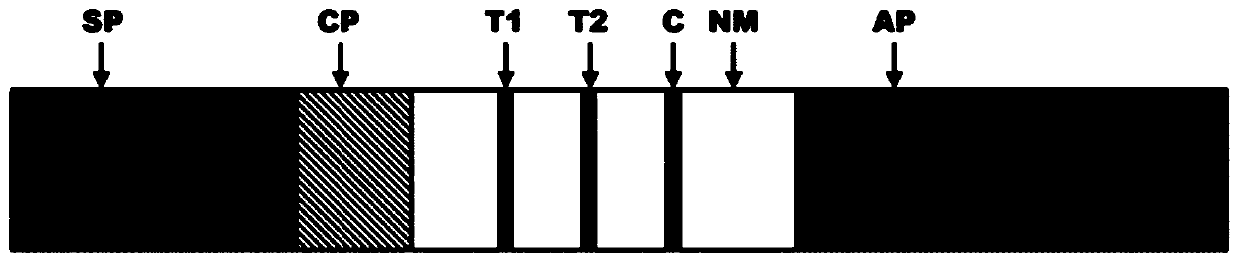
[0030] See attached Figure 1A , wherein, SP: sample pad CP: binding pad NM: nitrocellulose membrane detection area AP: absorbent pad.
[0031] (1) Preparation of nitrocellulose membrane detection area:
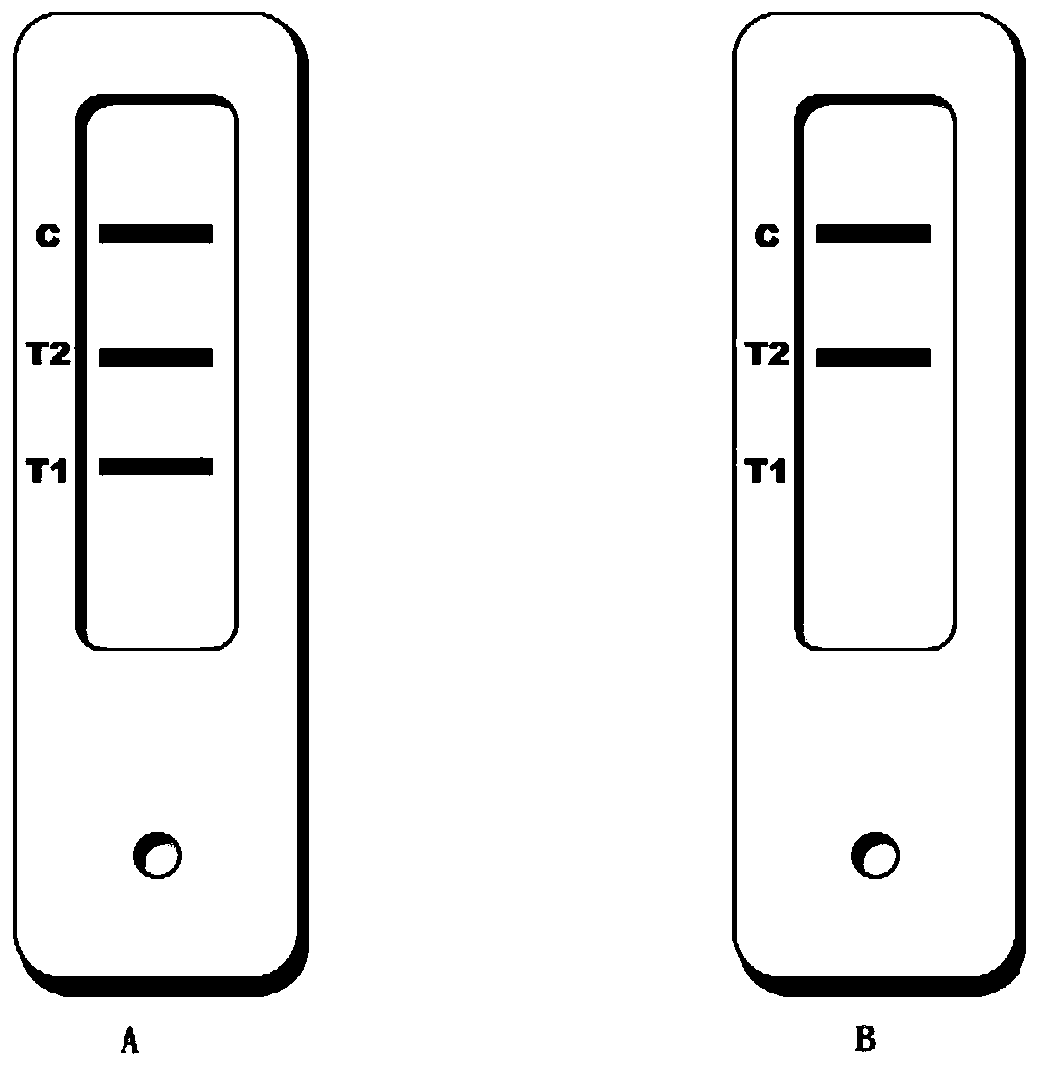
[0032] The nitrocellulose detection membrane was cut into long strips, placed on the platform of the dot-spraying apparatus, and digoxinumab, FITC monoclonal antibody and biotin-BSA were sprayed on the detection membrane respectively to form T1, T2 and C lines. Dry at 42°C for 30 minutes or dry naturally at room temperature.
[0033] (2) Preparation of binding pad:
[0034] Cut the glass wool into long strips, place it on the platform of the dot sprayer, take the nanoparticle markers, add a certain volume of pretreatment solution, spray dots on the glass wool, and dry at 50°C for 30 minutes.
[0035] (3) Preparation of sample pad:
[0036] Cut glass wool into long strips, soak with a certain...
Embodiment 2
[0039] Embodiment 2: sulfite solution treatment DNA
[0040] (1) Use a commercial DNA extraction kit or other appropriate methods to extract human genomic DNA;
[0041](2) Take 10 μL of DNA solution (100ng-2μg) and 90 μL of sulfite solution, mix well, centrifuge briefly, and place in a PCR instrument for reaction;
[0042] (3) The reaction conditions are as follows:
[0043] The first stage: 95°C for 5 minutes, 1 cycle;
[0044] The second stage: 95°C 30S, 70°C 10min, 15 cycles;
[0045] (4) Purify the DNA using a commercial DNA purification kit or other suitable methods.
Embodiment 3
[0046] Example 3: Methylation-dependent restriction endonuclease treatment of DNA
[0047] (1) Use a commercial DNA extraction kit or other appropriate methods to extract human genomic DNA;
[0048] (2) Take 10 μL DNA solution (100ng-2μg), 5 μL 10x reaction buffer, 2U methylation-dependent restriction endonuclease and 35 μL deionized water, mix well, centrifuge briefly, and place in a PCR instrument for reaction;
[0049] (3) The reaction conditions are as follows:
[0050] The first stage: 30 ℃ 1hr, 1 cycle,;
[0051] The second stage: 15min at 70°C, 1 cycle.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com