Transcription factor binding site prediction algorithm and device across transcription factors
A transcription factor and binding site technology, applied in the field of bioinformatics, can solve problems that cannot be used to predict transcription factor binding sites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
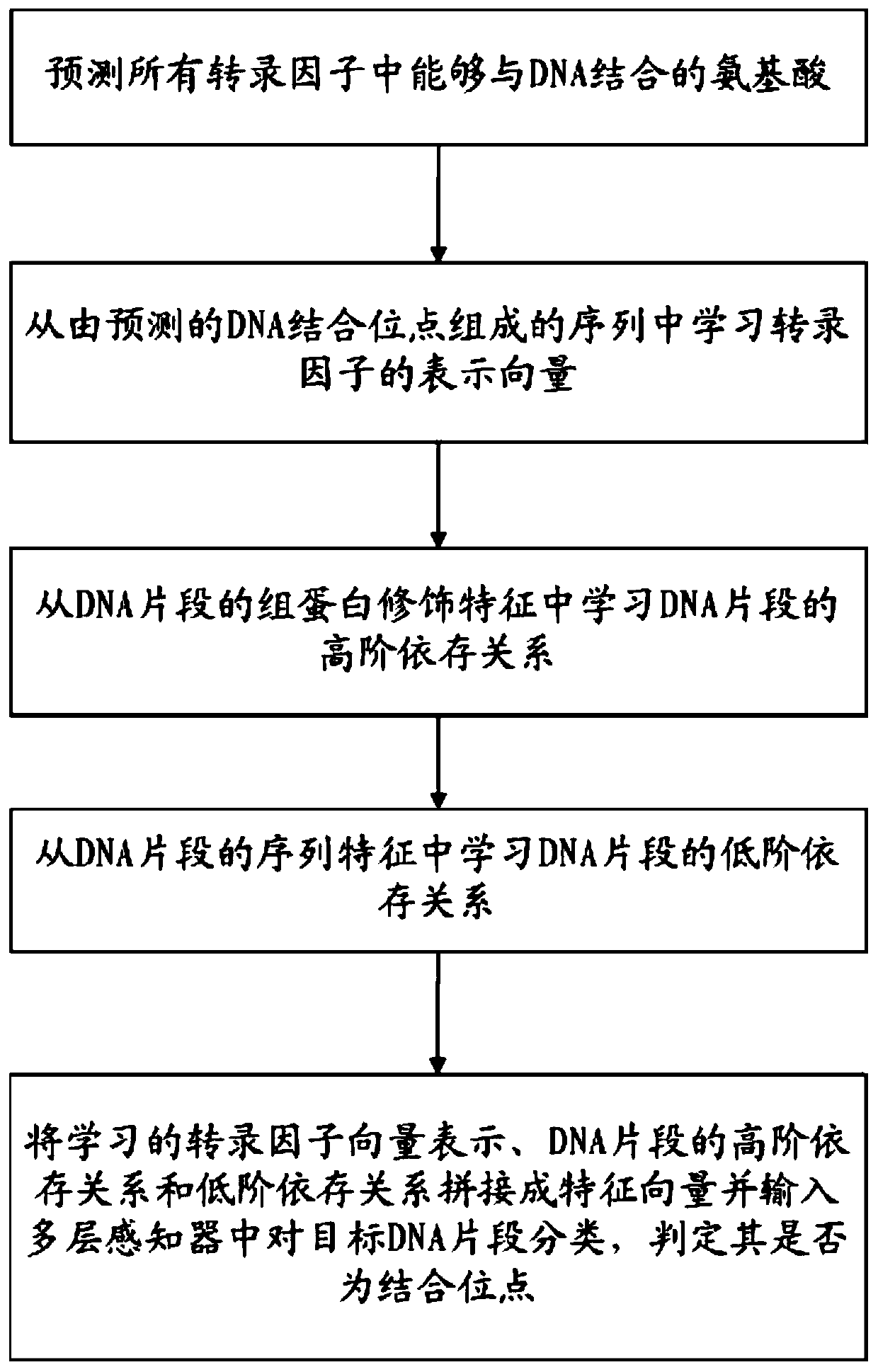
[0035] refer to figure 1 , figure 2 , Embodiment 1 of the present invention provides a binding site prediction method across transcription factors, the main steps are:
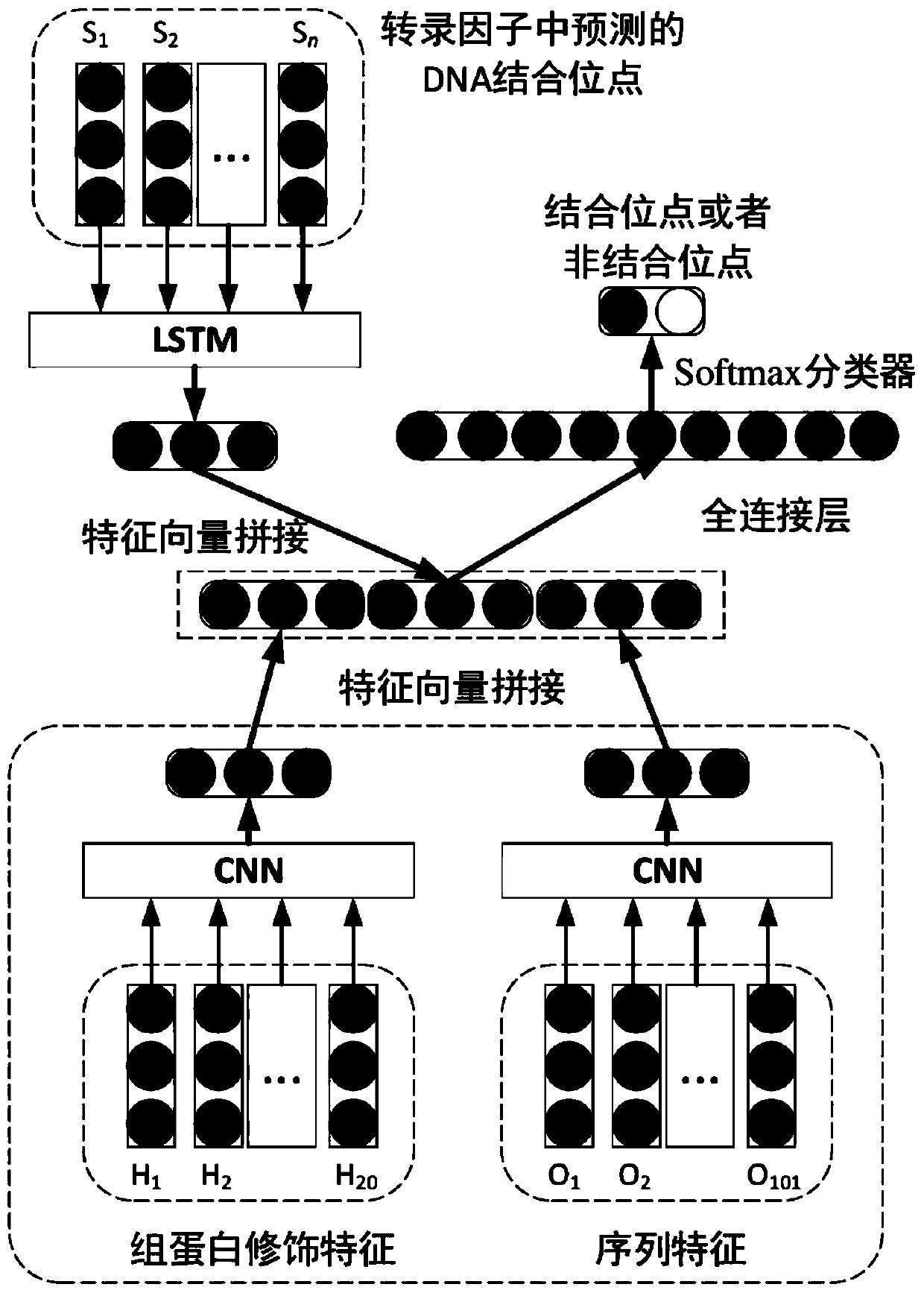
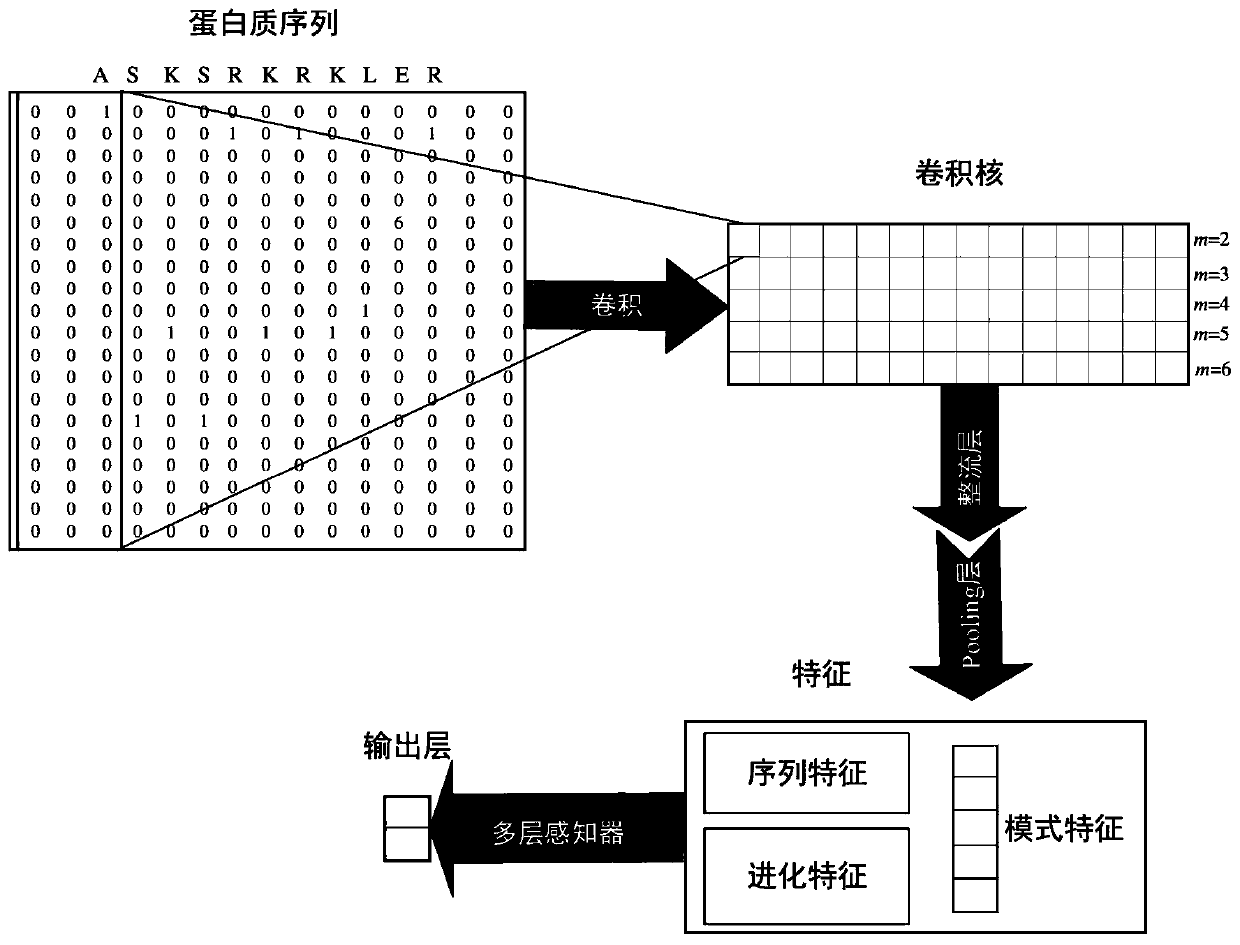
[0036] 1. Use the convolutional neural network model to predict the amino acids that can bind to DNA in all transcription factors, which are called DNA binding sites. The predicted DNA binding sites are mainly used to measure the labeled data of different transcription factors in the training process of the target transcription factor model contribution in .
[0037] Second, we use a long-short memory network model (LSTM) to learn representation vectors for transcription factors from sequences consisting of predicted DNA-binding sites.
[0038] 3. Using a convolutional neural network model to learn the high-order dependencies of DNA fragments from their histone modification signatures.
[0039] Fourth, use the convolutional neural network model to learn the low-order dependencies of DNA fragments from the ...
Embodiment 2
[0046] Embodiment 2 of the present invention provides a device for predicting binding sites across transcription factors, which mainly includes the following modules:
[0047] The DNA binding site prediction module is used to use the convolutional neural network model to predict the amino acids that can bind to DNA in all transcription factors, called DNA binding sites. The predicted DNA binding sites are mainly used to measure the labeling of different transcription factors Contribution of data during training of target transcription factor model.
[0048] A module for learning representation vectors for transcription factors, for learning representation vectors for transcription factors from a sequence consisting of predicted DNA binding sites using a long-short memory network model (LSTM).
[0049] A module for learning high-order dependencies of DNA fragments for learning high-order dependencies of DNA fragments from histone modification signatures of DNA fragments using a c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com