x-str fluorescence amplification system, kit and application
A technology of X-STR and amplification system, which is applied in the field of X-STR fluorescence amplification system to achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
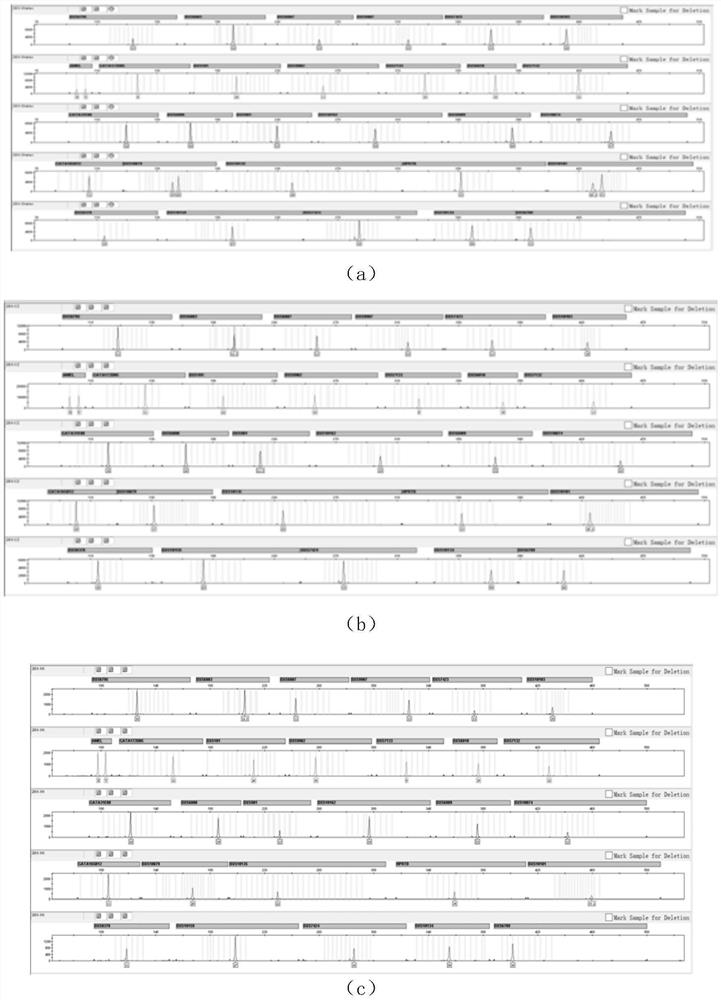
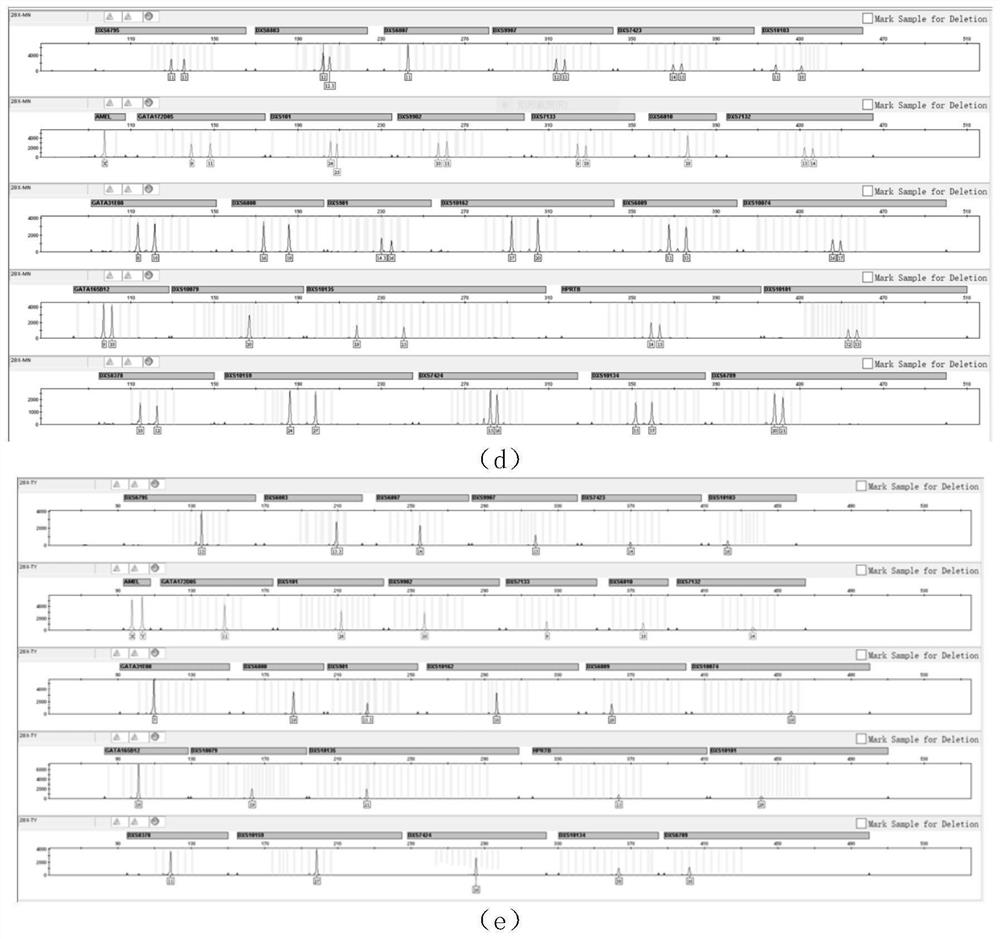
[0026] In order to make the object, technical solution and advantages of the present invention more clear, the present invention will be further described in detail below in conjunction with the examples. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
[0027] 1) Screening of X-STR loci:
[0028] The statistics of the genetic data of the 28 X-STRs screened by the present invention are shown in the results (Table 1).
[0029] Table 1: Statistical table of genetic data of 28 X-STRs used in the present invention
[0030]
[0031]
[0032] 2) Primer design:
[0033] The primers are designed using software such as Prime Premier5 and NCBI Blast. When designing primers, it should be ensured that the Tm value of each primer is within the range of (60±2)°C, and the amplification efficiency is close to that of the amplification product of each pair of primers. different eno...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com