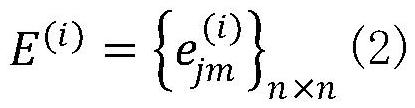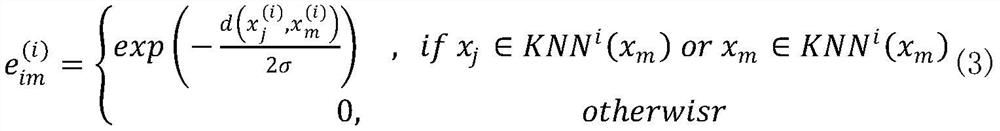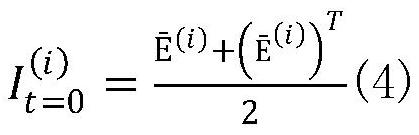Single cell integrated clustering method based on subspace randomization
A clustering method, single-cell technology, applied in the field of data mining in bioinformatics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0040] The present invention is based on subspace randomization single cell integrated clustering method. Specific embodiments of the present invention are described below. Those skilled in the art should understand that these embodiments are only used to explain the technical principles of the present invention, and are not intended to limit the scope of evidence collection of the present invention.
[0041] Step 1: From the GEO database, download 8 types of single-cell sequencing data, including Buettner (single-cell RNA sequencing of mouse embryonic stem cells), Deng (single-cell sequencing of mouse early development), Kolodziejczyk, Chu (single-cell sequencing of human embryonic stem cells Cell Sequencing), Usoskin (single-cell RNA-sequencing data of mouse sensory neurons), Zeisel (large-scale single-cell RNA-sequencing of mouse somatosensory cortex and hippocampal CA1 region). As the initial input data, in order to eliminate the influence of different dimensions, we norm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com