Design and synthesis method of nucleic acid coding probes for high-throughput sequencing
A coding and nucleic acid technology, which is applied in the design and synthesis of nucleic acid coding probes, can solve the problems of complex sequence modification process, difficulty in corresponding coding sequences to cells, coding sequences only to separate cells, etc., and achieve convenient, fast, accurate and accurate decoding Reliable sequencing results, reducing the troublesome effect of bioinformatics analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1, Design of High-throughput Sequencing Nucleic Acid Encoding Probe
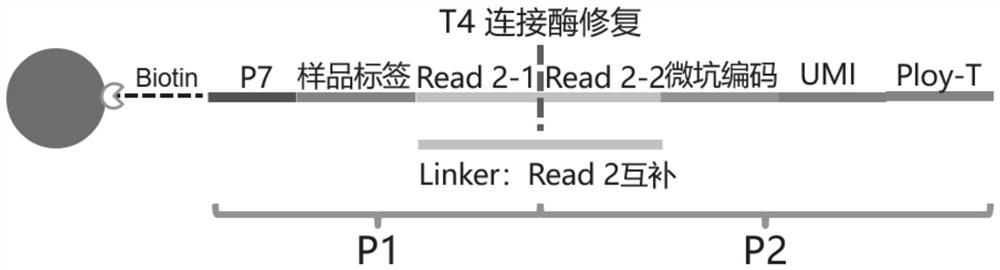
[0072] 1. Schematic diagram of the structure of high-throughput sequencing nucleic acid-encoded probes figure 1 , including the following components from the 5' end:
[0073] 1) Sequencing Adapters
[0074] The embodiment of the present invention uses Illumina's next-generation sequencing, therefore, the sequencing adapter is a P7 sequencing adapter (5'-CAAGCAGAAGACGGCATACGAGAT-3', sequence 1).
[0075] A space sequence is connected to the 5' end of the sequencing adapter (in the embodiment of the present invention, the space sequence is TTTTTTT, generally 5-10 Ts are appropriate), and the first base T of the space sequence is subjected to 5 The biotin modification at the 'end, the purpose of this space sequence is to provide a space for the connection of the probe to the streptavidin-modified magnetic beads during capture, so as to facilitate the connection of primers in the subsequent PCR ...
Embodiment 2
[0099] Embodiment 2, the establishment of nucleic acid coding probe synthesis method
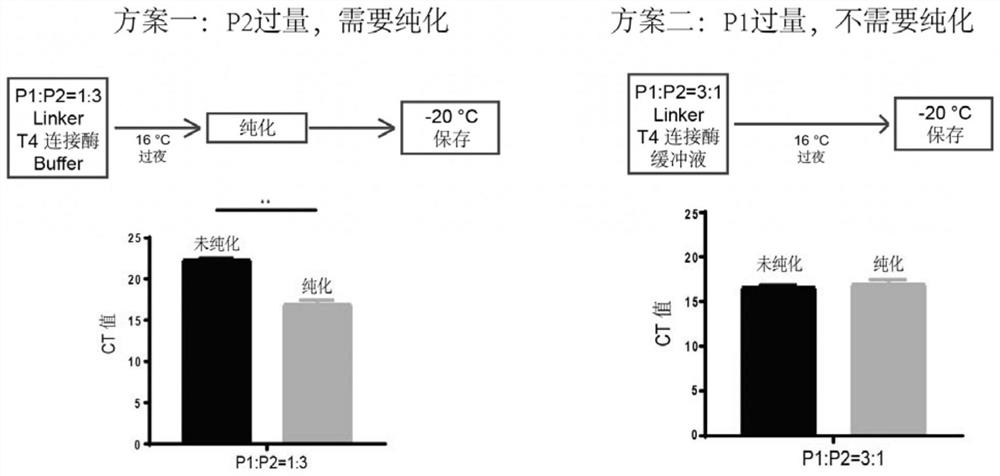
[0100] After determining the structural design of the encoded probe and the resolution and synthesis scheme, it is necessary to explore and optimize the specific synthesis method and conditions of the encoded probe. The schematic diagram of T4 DNA ligase synthesis of nucleic acid-encoded probes is as follows figure 2 As shown, by splitting the long sequence of the complete nucleic acid-encoded probe into two short sequences (P1 and P2) to be synthesized separately, the two parts can be synthesized into a complete capture probe under the action of T4 DNA ligase.
[0101] Take the following nucleic acid-encoded probe as an example (without UMI sequence, this is because sanger sequencing needs to determine the sequence, and the verification test does not need to add UMI):
[0102] TTTTTTTCAAGCAGAAGACGGCATACGAGAT CGTGAT GTGACTGGAGTTCAGACG(P1)TGTGCTCTTCCGATCTCGACACGGTTTGGGCCNNNNNNNNNNTTTTTTTT...
Embodiment 3
[0134] Example 3, Design of Nucleic Acid Encoded Probe and Experimental Verification of Synthesis Scheme
[0135] 1. Design of nucleic acid-encoded probes
[0136] According to the scheme of embodiment 1 one, design and synthesize the nucleic acid coded probe:
[0137]The nucleic acid-encoded probe is obtained by connecting P1 in Table 1 to P2 in Table 2, and the last base of p1 is adjacent to the first base of p2.
[0138] For example: P1-A in Table 1 is respectively connected to P2-1 to P2-96 in Table 2 to form 96 nucleic acid-encoded probes;
[0139] P1-B in Table 1 is respectively connected to P2-1 to P2-96 in Table 2 to form 96 nucleic acid-encoded probes;
[0140] P1-C in Table 2 are respectively connected to P2-1 to P2-96 in Table 2 to form 96 nucleic acid-encoded probes;
[0141] P1-D in Table 3 are respectively connected to P2-1 to P2-96 in Table 2 to form 96 nucleic acid-encoded probes;
[0142] In the embodiment of the present invention, a total of 384 nucleic a...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap